Phylogeographic differentiation of mitochondrial DNA in Han Chinese
- PMID: 11836649
- PMCID: PMC384943
- DOI: 10.1086/338999
Phylogeographic differentiation of mitochondrial DNA in Han Chinese
Abstract
To characterize the mitochondrial DNA (mtDNA) variation in Han Chinese from several provinces of China, we have sequenced the two hypervariable segments of the control region and the segment spanning nucleotide positions 10171-10659 of the coding region, and we have identified a number of specific coding-region mutations by direct sequencing or restriction-fragment-length-polymorphism tests. This allows us to define new haplogroups (clades of the mtDNA phylogeny) and to dissect the Han mtDNA pool on a phylogenetic basis, which is a prerequisite for any fine-grained phylogeographic analysis, the interpretation of ancient mtDNA, or future complete mtDNA sequencing efforts. Some of the haplogroups under study differ considerably in frequencies across different provinces. The southernmost provinces show more pronounced contrasts in their regional Han mtDNA pools than the central and northern provinces. These and other features of the geographical distribution of the mtDNA haplogroups observed in the Han Chinese make an initial Paleolithic colonization from south to north plausible but would suggest subsequent migration events in China that mainly proceeded from north to south and east to west. Lumping together all regional Han mtDNA pools into one fictive general mtDNA pool or choosing one or two regional Han populations to represent all Han Chinese is inappropriate for prehistoric considerations as well as for forensic purposes or medical disease studies.
Figures
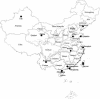
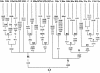

Similar articles
-
Analysis of mitochondrial DNA polymorphisms in Guangdong Han Chinese.Forensic Sci Int Genet. 2008 Mar;2(2):150-3. doi: 10.1016/j.fsigen.2007.10.122. Forensic Sci Int Genet. 2008. PMID: 19083810
-
Population data of mitochondrial DNA HVS-I and HVS-II sequences for 208 Henan Han Chinese.Leg Med (Tokyo). 2015 Jul;17(4):287-94. doi: 10.1016/j.legalmed.2015.02.003. Epub 2015 Feb 24. Leg Med (Tokyo). 2015. PMID: 25759193
-
Phylogeographic analysis of mtDNA variation in four ethnic populations from Yunnan Province: new data and a reappraisal.J Hum Genet. 2002;47(6):311-8. doi: 10.1007/s100380200042. J Hum Genet. 2002. PMID: 12111379
-
Phylogeographic analysis of mitochondrial DNA in northern Asian populations.Am J Hum Genet. 2007 Nov;81(5):1025-41. doi: 10.1086/522933. Epub 2007 Oct 1. Am J Hum Genet. 2007. PMID: 17924343 Free PMC article.
-
Ancient DNA reveals genetic connections between early Di-Qiang and Han Chinese.BMC Evol Biol. 2017 Dec 4;17(1):239. doi: 10.1186/s12862-017-1082-0. BMC Evol Biol. 2017. PMID: 29202706 Free PMC article.
Cited by
-
Genomic insights into the genetic structure and population history of Mongolians in Liaoning Province.Front Genet. 2022 Oct 12;13:947758. doi: 10.3389/fgene.2022.947758. eCollection 2022. Front Genet. 2022. PMID: 36313460 Free PMC article.
-
Ancient inland human dispersals from Myanmar into interior East Asia since the Late Pleistocene.Sci Rep. 2015 Mar 26;5:9473. doi: 10.1038/srep09473. Sci Rep. 2015. PMID: 25826227 Free PMC article.
-
Hypervariable region polymorphism of mtDNA of recurrent oral ulceration in Chinese.PLoS One. 2012;7(9):e45359. doi: 10.1371/journal.pone.0045359. Epub 2012 Sep 19. PLoS One. 2012. PMID: 23028959 Free PMC article.
-
Mitochondrial DNA haplogroups and short-term neurological outcomes of ischemic stroke.Sci Rep. 2015 May 20;5:9864. doi: 10.1038/srep09864. Sci Rep. 2015. PMID: 25993529 Free PMC article.
-
Updating phylogeny of mitochondrial DNA macrohaplogroup m in India: dispersal of modern human in South Asian corridor.PLoS One. 2009 Oct 13;4(10):e7447. doi: 10.1371/journal.pone.0007447. PLoS One. 2009. PMID: 19823670 Free PMC article.
References
Electronic-Database Information
-
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for mtDNA control region data; accession numbers AY052834–AY053358)
References
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
-
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 - PubMed
-
- Bandelt H-J, Macaulay V, Richards M (2000) Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol Phylogenet Evol 16:8–28 - PubMed
-
- Bandelt H-J, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

