-
PDF
- Split View
-
Views
-
Cite
Cite
Svenja Heesch, Martha Serrano‐Serrano, Josué Barrera‐Redondo, Rémy Luthringer, Akira F. Peters, Christophe Destombe, J. Mark Cock, Myriam Valero, Denis Roze, Nicolas Salamin, Susana M. Coelho, Evolution of life cycles and reproductive traits: Insights from the brown algae, Journal of Evolutionary Biology, Volume 34, Issue 7, 1 July 2021, Pages 992–1009, https://doi.org/10.1111/jeb.13880
Close - Share Icon Share
Abstract
A vast diversity of types of life cycles exists in nature, and several theories have been advanced to explain how this diversity has evolved and how each type of life cycle is retained over evolutionary time. Here, we exploited the diversity of life cycles and reproductive traits of the brown algae (Phaeophyceae) to test several hypotheses on the evolution of life cycles. We investigated the evolutionary dynamics of four life‐history traits: life cycle, sexual system, level of gamete dimorphism and gamete parthenogenetic capacity. We assigned states to up to 77 representative species of the taxonomic diversity of the brown algal group, in a multi‐gene phylogeny. We used maximum likelihood and Bayesian analyses of correlated evolution, while taking the phylogeny into account, to test for correlations between traits and to investigate the chronological sequence of trait acquisition. Our analyses are consistent with the prediction that diploid growth evolves when sexual reproduction is preferred over asexual reproduction, possibly because it allows the complementation of deleterious mutations. We also found that haploid sex determination is ancestral in relation to diploid sex determination. However, our results could not address whether increased zygotic and diploid growth are associated with increased sexual dimorphism. Our analyses suggest that in the brown algae, isogamous species evolved from anisogamous ancestors, contrary to the commonly reported pattern where evolution proceeds from isogamy to anisogamy.
Abstract
INTRODUCTION
The life cycle of an organism is one of its most fundamental features and influences the evolution of a variety of traits, including mode of reproduction, developmental processes, mode of dispersal, adaptation to local environment and ecological success. A wide variety of different life cycles are found within eukaryotes, and one of the great challenges of evolutionary biology is to understand how this diversity has evolved, and how each type of life cycle is retained within a lineage at evolutionary timescales (Cock et al., 2014; Mable & Otto, 1998; Otto & Gerstein, 2008; Valero et al., 1992).
The sexual life cycle of eukaryotes involves the fusion of two gametes to form a zygote, followed by meiosis. Such life cycles can be divided into three main types: haplontic, where only the haploid phase undergoes mitosis; diplontic, where only the diploid phase undergoes mitosis; and diplohaplontic (or haploid‐diploid), where both phases undergo mitosis (Coelho et al., 2007; Otto & Gerstein, 2008; Valero et al., 1992). In photosynthetic organisms, multicellular haploid phases are usually termed gametophytes since they produce gametes, and multicellular diploid phases are called sporophytes since they produce haploid spores. Diplohaplontic life cycles may be iso‐ or heteromorphic. For the latter, the dominant phase may be haploid (such as in mosses) or diploid (such as in vascular plants and kelps). Asymmetry in terms of the length and complexity of the haploid and diploid phases can be very strong (Lipinska et al., 2019) and can eventually lead to transitions towards diplontic or haplontic life cycles.
The structure of an organism's life cycle also has important consequences for the evolution of its sex determination system (Coelho et al., 2018). Haploid sex determination is common in diplohaplontic lineages such as in brown algae (Phaeophyceae), where gametophytes can either be monoicous or dioicous (Table 1). In gymnosperms and angiosperms, sex is determined in the diploid phase and the organism may be monoecious if a single individual produces female and male gametes or dioecious if male and female gametes are produced by two different individuals. Correlations between the type of sexual system and life‐history features such as gamete size, antheridium number, ploidy level and diversification rate are relatively well studied in angiosperms and mosses (Goldberg et al., 2017; Villarreal & Renner, 2013) but studies of other eukaryotic groups are virtually inexistent.
Description of the traits studied, categories and discrete states. Note that some of the discrete traits were also treated as continuous traits (male gamete size for instance)
| Trait | Category | States | Description |
| Life cycle | Diplohaplontic | Diplohaplontic haploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant gametophyte (haploid) generation |
| Diplohaplontic haploid = diploid | Life cycle with both haploid and diploid mitosis, with equal dominance of gametophyte and sporophyte generations | ||
| Diplohaplontic diploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant sporophyte (diploid) generation | ||
| Diplontic | Diplontic | Life cycle with no haploid mitosis, the haploid phase is limited to gametes | |
| Sexual system | Haploid sex determination | Monoicous | Haploid phase sex determination, where both gamete types are produced by the same haploid gametophyte |
| Dioicous | Haploid phase sex or mating type determination, with genetically distinct gametophytes corresponding to each sex | ||
| Diploid sex determination | Dioecious | Diploid phase sex determination, with genetically distinct sporophytes corresponding to each sex | |
| Monoecious | Diploid phase sex determination, where both male and female organs are produced by the same diploid sporophyte | ||
| Gamete size | Female gamete with flagella | Isogamous b | Male and female gametes with no noticeable size difference (but different behaviour/physiology) |
| Anisogamous | Male and female gametes of clearly different size, both with flagella | ||
| Female gamete without flagella | Oogamous | Female gamete much larger and lacking a flagellum | |
| Parthenogenesis | No parthenogenesis | No parthenogenesis | No parthenogenesis capacity in either gamete |
| Parthenogenesis | Female gametes only | Only female gametes capable of parthenogenesis | |
| Male and female gametes | Male and female gametes capable of parthenogenesis |
| Trait | Category | States | Description |
| Life cycle | Diplohaplontic | Diplohaplontic haploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant gametophyte (haploid) generation |
| Diplohaplontic haploid = diploid | Life cycle with both haploid and diploid mitosis, with equal dominance of gametophyte and sporophyte generations | ||
| Diplohaplontic diploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant sporophyte (diploid) generation | ||
| Diplontic | Diplontic | Life cycle with no haploid mitosis, the haploid phase is limited to gametes | |
| Sexual system | Haploid sex determination | Monoicous | Haploid phase sex determination, where both gamete types are produced by the same haploid gametophyte |
| Dioicous | Haploid phase sex or mating type determination, with genetically distinct gametophytes corresponding to each sex | ||
| Diploid sex determination | Dioecious | Diploid phase sex determination, with genetically distinct sporophytes corresponding to each sex | |
| Monoecious | Diploid phase sex determination, where both male and female organs are produced by the same diploid sporophyte | ||
| Gamete size | Female gamete with flagella | Isogamous b | Male and female gametes with no noticeable size difference (but different behaviour/physiology) |
| Anisogamous | Male and female gametes of clearly different size, both with flagella | ||
| Female gamete without flagella | Oogamous | Female gamete much larger and lacking a flagellum | |
| Parthenogenesis | No parthenogenesis | No parthenogenesis | No parthenogenesis capacity in either gamete |
| Parthenogenesis | Female gametes only | Only female gametes capable of parthenogenesis | |
| Male and female gametes | Male and female gametes capable of parthenogenesis |
The term ‘dominant’ is defined here as the generation that presents larger size and higher complexity in terms of morphology (number of different cell types, number of tissues and organs).
For simplicity, we code as ‘isogamous’ algae that have almost imperceptible size differences between male and female gametes, but note that in the brown algae there is always an asymmetry (at least in terms of physiology and behaviour) between male and female gametes.
Description of the traits studied, categories and discrete states. Note that some of the discrete traits were also treated as continuous traits (male gamete size for instance)
| Trait | Category | States | Description |
| Life cycle | Diplohaplontic | Diplohaplontic haploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant gametophyte (haploid) generation |
| Diplohaplontic haploid = diploid | Life cycle with both haploid and diploid mitosis, with equal dominance of gametophyte and sporophyte generations | ||
| Diplohaplontic diploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant sporophyte (diploid) generation | ||
| Diplontic | Diplontic | Life cycle with no haploid mitosis, the haploid phase is limited to gametes | |
| Sexual system | Haploid sex determination | Monoicous | Haploid phase sex determination, where both gamete types are produced by the same haploid gametophyte |
| Dioicous | Haploid phase sex or mating type determination, with genetically distinct gametophytes corresponding to each sex | ||
| Diploid sex determination | Dioecious | Diploid phase sex determination, with genetically distinct sporophytes corresponding to each sex | |
| Monoecious | Diploid phase sex determination, where both male and female organs are produced by the same diploid sporophyte | ||
| Gamete size | Female gamete with flagella | Isogamous b | Male and female gametes with no noticeable size difference (but different behaviour/physiology) |
| Anisogamous | Male and female gametes of clearly different size, both with flagella | ||
| Female gamete without flagella | Oogamous | Female gamete much larger and lacking a flagellum | |
| Parthenogenesis | No parthenogenesis | No parthenogenesis | No parthenogenesis capacity in either gamete |
| Parthenogenesis | Female gametes only | Only female gametes capable of parthenogenesis | |
| Male and female gametes | Male and female gametes capable of parthenogenesis |
| Trait | Category | States | Description |
| Life cycle | Diplohaplontic | Diplohaplontic haploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant gametophyte (haploid) generation |
| Diplohaplontic haploid = diploid | Life cycle with both haploid and diploid mitosis, with equal dominance of gametophyte and sporophyte generations | ||
| Diplohaplontic diploid‐dominant a | Life cycle with both haploid and diploid mitosis, with dominant sporophyte (diploid) generation | ||
| Diplontic | Diplontic | Life cycle with no haploid mitosis, the haploid phase is limited to gametes | |
| Sexual system | Haploid sex determination | Monoicous | Haploid phase sex determination, where both gamete types are produced by the same haploid gametophyte |
| Dioicous | Haploid phase sex or mating type determination, with genetically distinct gametophytes corresponding to each sex | ||
| Diploid sex determination | Dioecious | Diploid phase sex determination, with genetically distinct sporophytes corresponding to each sex | |
| Monoecious | Diploid phase sex determination, where both male and female organs are produced by the same diploid sporophyte | ||
| Gamete size | Female gamete with flagella | Isogamous b | Male and female gametes with no noticeable size difference (but different behaviour/physiology) |
| Anisogamous | Male and female gametes of clearly different size, both with flagella | ||
| Female gamete without flagella | Oogamous | Female gamete much larger and lacking a flagellum | |
| Parthenogenesis | No parthenogenesis | No parthenogenesis | No parthenogenesis capacity in either gamete |
| Parthenogenesis | Female gametes only | Only female gametes capable of parthenogenesis | |
| Male and female gametes | Male and female gametes capable of parthenogenesis |
The term ‘dominant’ is defined here as the generation that presents larger size and higher complexity in terms of morphology (number of different cell types, number of tissues and organs).
For simplicity, we code as ‘isogamous’ algae that have almost imperceptible size differences between male and female gametes, but note that in the brown algae there is always an asymmetry (at least in terms of physiology and behaviour) between male and female gametes.
One important feature of sexual life cycles in eukaryotes is the degree of similarity between male and female gametes. This ‘gamete dimorphism’ is a continuous trait, and a number of models have been proposed to explain how anisogamous organisms could evolve from an isogamous ancestor (Hoekstra, 1980; Randerson & Hurst, 2001). The evolution of anisogamy establishes the fundamental basis for maleness and femaleness and leads to an asymmetry in resource allocation to the offspring, leading in many cases to sexual selection (Billiard et al., 2011). Anisogamy and oogamy have arisen repeatedly across the eukaryotes, and these systems are thought to be derived from simpler isogamous mating systems, either due to disruptive selection generated by a trade‐off between the number of offspring produced and offspring survival (Bulmer & Parker, 2002; Parker, 1978), to selection to maximize the rate of gamete encounter (Dusenbery, 2000; Togashi et al., 2012), or as a mechanism to reduce cytoplasmic conflicts (Hurst & Hamilton, 1992; Hutson & Law, 1993).
Differences in gamete size in anisogamous and oogamous species may influence other reproductive characteristics, such as the capacity of undergoing asexual reproduction through parthenogenesis (Billiard et al., 2011; Hoekstra, 1980). In animals and land plants, parthenogenesis has been mostly described for females only (Dawley & Bogart, 1989), but in organisms with moderate levels of gamete dimorphism such as some brown algae, development from both male and female gametes in the absence of fertilization is quite common, at least under laboratory conditions (Bothwell et al., 2010; Mignerot et al., 2019; Oppliger et al., 2007).
The different types of life cycles have evolved independently and repeatedly in different eukaryotic groups, and this is also the case for the types of sexual systems. Testing evolutionary hypotheses regarding the causes and consequences of life‐history trait diversity requires data from multiple species placed in a phylogenetic context. Such comparative studies have been hampered by a lack of accessible data regarding life cycles, sexual systems and sex determination mechanisms across the eukaryotic tree of life, and most specifically in groups outside animals and land plants. Although knowledge has been recently growing in Chloroplastida, with studies extending to bryophytes and volvocine algae (Hanschen et al., 2018; Villarreal & Renner, 2013), we still lack views on other eukaryotic groups, that should help us understand the general principles underlying the evolution of these traits.
The brown algae represent a fascinating group to study the evolution of life cycles and reproductive traits, since they exhibit a remarkable range of life cycles and sexual traits (Bell, 1997; Clayton, 1988; Figure 1). In 1997, Bell used the diversity of life cycles within the brown algae to test hypotheses on the evolution of life cycles; in particular, whether evolution generally proceeds towards an increase of the diploid phase at the expense of the haploid phase (Clayton, 1988), and whether a positive association between a prolonged haploid phase and the rate of inbreeding (as predicted by theories based on the effect of deleterious alleles; Otto & Marks, 1996) is observed (using gametophyte monoicy as a proxy for inbreeding by assuming that gametophytic selfing may occur). However, his study was based on a phylogenetic tree including only 14 species, and evolutionary relationships between brown algal orders were at the time poorly resolved, making it difficult to test his assumptions.
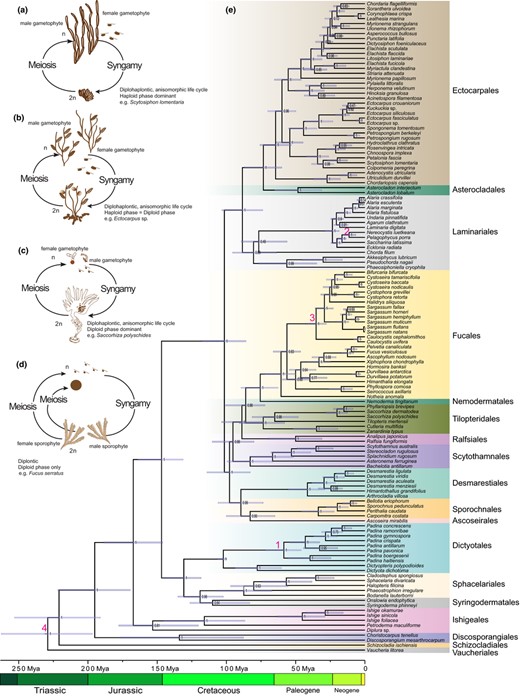
Schematic illustration of sexual life cycles of representative brown algae and brown algal phylogenetic tree. (a) Scytosiphon lomentaria (Lyngbye) Link: diplohaplontic, heteromorphic life cycle, haploid dominant; near‐isogametes (b) Ectocarpus sp.: diplohaplontic, isomorphic life cycle, with similar dominance in haploid and diploid phases (H=D); near‐isogametes; (c) Saccorhiza polyschides (Lightfoot) Batters: diplohaplontic, heteromorphic life cycle, with diploid dominance (D>>H); oogamous; (d) Fucus serratus L.: diplontic life cycle, only diploid phase; oogamous. H = haploid phase; D = diploid phase. (e) Phylogenetic tree using Bayesian analyses in BEAST. Node numbers indicate the posterior Bayesian support, node bars represent the 95% HPD (Highest Posterior Density) for the divergence times. Node calibration times are indicated by numbers 1–4
In this study, we exploited a well‐resolved phylogeny of 91 species of brown algae (Silberfeld et al., 2010, 2014) and extended it to 131 species, containing representatives from 16 of the 20 brown algae orders, which are roughly composed of ~300 genera and ~2,000 species (Silberfeld et al., 2014), in order to understand how life cycles and reproductive traits evolved across Phaeophyceae. We performed an extensive literature review to recover information for life cycle and reproductive traits across the brown algae. We could recover information for a maximum of 77 species, representative of most orders of brown algae (Dataset S1). We estimated ancestral states for each of the traits, as well as the number of transitions between states and their relative timing, and assessed possible correlations between the life cycle and reproductive traits. These analyses have allowed us to describe the evolution of life cycles and reproductive traits across the brown algal phylogeny and to test a number of long‐standing hypotheses about the evolution of life cycles and reproductive traits such as (1) the possibility that diploid growth evolved alongside a higher tendency towards sexual reproduction (Otto & Goldstein, 1992; Otto & Marks, 1996), (2) whether increased zygotic and diploid growth are associated with increased sexual dimorphism (Bell, 1994), (3) whether haploid sex determination is ancestral in relation to diploid sex determination (4) and whether anisogamous species evolved from isogamous ancestors (Bell, 1978; Parker et al., 1972). We also tested additional hypotheses, including the possibility that gamete size influences the capacity for asexual reproduction through parthenogenesis (Luthringer et al., 2014), and we discuss the macroevolutionary dynamics of transitions between sexual systems in the brown algae.
METHODS
Molecular data
Multiple sequence alignments were performed for 131 brown algae species, based on the nucleotide data published by Silberfeld et al., (2010, 2014), corresponding to five mitochondrial genes (atp9: mitochondrial ATP synthase subunit 9 gene, cox1 and cox4: Cytochrome c oxidase subunit 1 and 3 genes, nad1 and nad4: NADH dehydrogenase subunit 1 and 4), four chloroplast genes (rbcL: large subunit of plastid‐encoded ribulose‐1,5‐biphosphate carboxylase oxygenase gene, psaA: photosystem I P700 chlorophyll a apoprotein A1 gene, psbA: photosystem II protein D1 gene, and atpB: ATP synthase subunit b gene) and one nuclear gene (LSU: large subunit of 28S rRNA gene). To attribute trait states to each species, we replaced some entities, depending on the availability of life‐history information (i.e. kept the sequence data used to build the tree but used the data on life history from another close relative; Table 1). Accession numbers for the sequences of the species that were not included in Silberfeld et al. (2010, 2014) are in Table S1. No information was available about the life histories of the closest relatives of the Phaeophyceae, for example Phaeothamniophyceae, so we used Schizocladia and Vaucheria as outgroups, both heterokont genera from the classes Schizocladiophyceae and Xanthophyceae from which Vaucheria had available life cycle and reproductive trait information. The final species list used for the trait analysis, for which we had life cycle and reproductive trait information, comprised of 77 species, including the outgroup.
Phylogenetic reconstruction
We used the sequence data from the 131 brown algae species to infer a phylogenetic tree (Figure 1). All sequences were aligned using MAFFT (Katoh et al., 2009), and the best substitution models were estimated as GTR+G for three different gene partitions, corresponding to the nuclear, plastid and mitochondrial genes using the phymltest function in the ape R package (Paradis et al., 2004). The concatenated alignment (TreeBASE submission ID S28254) was used for Bayesian Inference with Beast v1.8.2 (Drummond et al., 2012). Each partition was unlinked for the substitution model. We used birth‐death with incomplete sampling as tree prior and four calibration nodes as described in Silberfeld et al. (2010) (see nodes 1 to 4, Figure 2). We used lognormal priors for two of the calibrations: Padina‐like clade 1, lognormal distribution (mean 5 Ma, sd 1, and lower boundary at 99.6 Ma); Nereocystis‐Pelagophycus clade 2: lognormal distribution (mean 20 Ma, sd 1, and lower boundary at 13 Ma), normal priors for the root (Phaeophyceae root age 4: normal distribution (u = 155, sd=30 Ma) and a normal distribution for the Sargassaceae node 3 (u = 60, sd=15, with lower boundary 13 Ma). We also included a prior to separate Phaeophyceae as a monophyletic clade. Finally, the MCMC was set to 50 million generations with a sampling every 1,000 generations and a subsequent burn‐in of 16% of the sampled trees. The posterior distribution was summarized using Treeannotator v1.7.0 (Drummond et al., 2012) to obtain a Common Ancestor Tree (Heled & Bouckaert, 2013; TreeBASE submission ID S28254). For the macroevolutionary analyses (see below), a set of 100 trees were sampled from the posterior distribution.
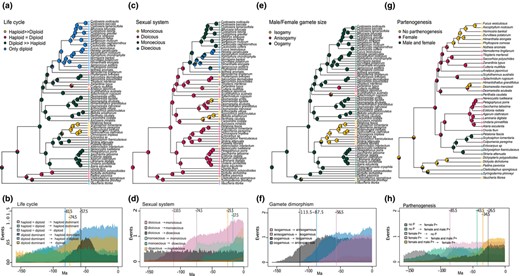
Maximum likelihood ancestral state reconstructions for the four‐brown algal life‐history traits. Pie charts and colours at each node represent the probabilities for each state. Colours at the tips represent the species states (a, c, e, g). Estimated number of transitions through time for the corresponding four life‐history traits (b, d, f, h). Coloured densities identify the mean number of events for each possible transition. Vertical lines and numbers denote the minimum age as the point in time, where at least one transition is recorded in 60% of the reconstructions. (a and b) Male and female gamete size; (c and d) sexual system, (e and f) type of life cycle, (g and h) parthenogenetic capacity. D>>H: diploid stage dominant; H>>D: haploid stage dominant; H=D: haploid and diploid stage with equal dominance
Life‐history traits
We estimated the ancestral state of each of the four main sexual traits: type of life cycle (haploid > diploid; haploid = diploid; haploid < diploid; diplont), type of sexual system (monoicous; dioicous; monoecious; dioecious), level of gamete dimorphism (isogamous; anisogamous; oogamous) and parthenogenetic capacity (no parthenogenesis; parthenogenesis in female gametes only; parthenogenesis in both male and female gametes). The traits were coded as discrete multi‐state characters (Table 2). Definitions of the life cycle and sexual terms used in this study are provided in Table 1. We separated the respective traits into seven additional characters. For example, we transferred ‘gamete size’ (iso‐, aniso‐, oogamous) into a continuous male gamete size trait. We furthermore recoded multi‐state traits into binary data for the correlation tests (see below), such as the ‘gamete dimorphism’, which was recorded by separating the absence (0 = oogamy) from presence (1 = iso‐ or anisogamy) of female flagellated gametes. We categorized an additional sexual system trait as ‘sexes occurring on the same thallus’ (0 = monoicous or monoecious) or ‘separate thalli’ (1 = dioicous or dioecious). The life cycle was simplified to the occurrence of a ‘dominant haploid phase’ (0 = haploid ≥diploid) versus dominance of the diploid phase (1 = haploid <diploid or diplontic), with dominance broadly meaning size of the adult individual. Finally, the occurrence of parthenogenesis was separated into two additional traits, absence (0) or presence (1) of male parthenogenesis, and absence of parthenogenesis (0) versus parthenogenesis occurring in at least one of the sexes (1), most commonly the female.
List of detailed life cycle and reproductive traits across the brown algal species
| Species | Gamete size ratio (F/M) | Gamete dimorphism | Male gamete size (um) | Isogamy versus anisogamy | Sexual system | HSD or DSD | Co‐sexual versus separate sexes | Type of life cycle | Generation dominance (simple) | Generation dominance | Parthenogenesis capacity | Both parthenogenesis | Parthenogenesis presence/absence | Male parthenogenesis |
| 0 = isogamous | 0 = isogamous | 0 = monoicous | 0 = haploid sex determination | 0 = sexes on same thallus | 0 = H/D with H > D | 0 = haploid‐dominant (or similar dominance) | 0 = H»D | 0 = no parthenogenesis | 0= none or 1 do | 0= no parthenogenesis | 0 = no male parthenogenesis | |||
| 1 = anisogamous | 1 = anisogamous and oogamous | 1 = dioicous | 1 = diploid sex determination | 1 = sexes on separate thalli | 1 = H/D with H = D | 1 = diploid‐dominant | 1 = H=D | 1 = female only | 1 = both do parthenogenesis | 1 = at least female does | 1 = male parthenogenesis | |||
| 2 = oogamous | 2 = monoecious | 2 = H/D with D > H | 2 = D»H | 2 = female and male | ||||||||||
| 3 = dioecious | 3= Diploid | 3 = Diplont (no H) | ||||||||||||
| Bifurcaria bifurcata | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira tamariscifolia | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira baccata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira nodicaulis | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora grevillei | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora retorta | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Halidrys siliquosa | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum fallax | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum muticum | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis cephalornithos | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis uvifera | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Phyllospora comosa | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Seirococcus axillaris | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Durvillaea potatorum | 12,4 | 2 | 3 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Himanthalia elongata | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Xiphophora chondrophylla | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Hormosira banksii | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Ascophyllum nodosum | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Fucus vesiculosus | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Pelvetia canaliculata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Notheia anomala | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Nemoderma tingitanum | 2,3 | 1 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Cutleria multifida | 5,2 | 1 | 5 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Zanardinia typus | 4,7 | 1 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Tilopteris mertensii | 7,5 | 2 | 8 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 1 | 0 | 1 | 0 |
| Saccorhiza polyschides | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Phyllariopsis brevipes | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | NA | NA | NA | NA |
| Saccorhiza dermatodea | 5,6 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Analipus japonicus | 1,2 | 1 | 7 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Ralfsia fungiformis | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ascoseira mirabilis | 1 | 0 | 8 | 0 | 2 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Agarum clathratum | 7,8 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Laminaria digitata | 2,9 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Alaria spp. | 3 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Undaria pinnatifida | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Ecklonia radiata | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Saccharina latissima | 3,6 | 1 | 9 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Nereocystis luetkeana | NA | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Pelagophycus porra | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Chorda filum | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Hydroclathrus clathratus | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Rosenvingea intricata | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Chnoospora implexa | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | NA | NA | NA | NA |
| Colpomenia peregrina | 1,5 | 1 | 4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Petalonia fascia | 1 | 0 | 7 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Scytosiphon lomentaria | 1,1 | 0 | 8 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Ectocarpus sp. | NA | 1 | 6 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 |
| Petrospongium berkeleyi | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Feldmannia mitchelliae | 1,7 | 1 | 11 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Pylaiella littoralis | 1 | 0 | NA | 0 | 1 | 0 | 1 | 1 | 0 | 1 | NA | NA | NA | NA |
| Elachista fucicola | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Chordaria linearis | NA | 0 | 8 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Dictyosiphon foeniculaceus | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Striaria attenuata | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Asterocladon interjectum | NA | 1 | 7 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Scytothamnus australis | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Splachnidium rugosum | 1,75 | 1 | 6 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Bachelotia antillarum | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Carpomitra costata | 5,5 | 2 | 5 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Perithalia caudata | 2,9 | 2 | 7 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Bellotia eriophorum | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Sporochnus pedunculatus | 7,5 | 2 | 4 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Arthrocladia villosa | 2,1 | 2 | 12 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia menziesii | 2,3 | 2 | Na | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Himantothallus grandifolius | 5 | 2 | 4 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Desmarestia aculeata | 6 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Desmarestia ligulata | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia viridis | 3,9 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Cladostephus spongiosus | 1 | 0 | NA | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Syringoderma phinneyi | 1 | 0 | NA | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Padina spp | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyopteris polypodioides | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyota dichotoma | 13,9 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Ishige okamurae | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Phaeosiphoniella cryophila | 15 | 2 | NA | 1 | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Schizocladia ischiensis | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Vaucheria litorea | 40 | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Species | Gamete size ratio (F/M) | Gamete dimorphism | Male gamete size (um) | Isogamy versus anisogamy | Sexual system | HSD or DSD | Co‐sexual versus separate sexes | Type of life cycle | Generation dominance (simple) | Generation dominance | Parthenogenesis capacity | Both parthenogenesis | Parthenogenesis presence/absence | Male parthenogenesis |
| 0 = isogamous | 0 = isogamous | 0 = monoicous | 0 = haploid sex determination | 0 = sexes on same thallus | 0 = H/D with H > D | 0 = haploid‐dominant (or similar dominance) | 0 = H»D | 0 = no parthenogenesis | 0= none or 1 do | 0= no parthenogenesis | 0 = no male parthenogenesis | |||
| 1 = anisogamous | 1 = anisogamous and oogamous | 1 = dioicous | 1 = diploid sex determination | 1 = sexes on separate thalli | 1 = H/D with H = D | 1 = diploid‐dominant | 1 = H=D | 1 = female only | 1 = both do parthenogenesis | 1 = at least female does | 1 = male parthenogenesis | |||
| 2 = oogamous | 2 = monoecious | 2 = H/D with D > H | 2 = D»H | 2 = female and male | ||||||||||
| 3 = dioecious | 3= Diploid | 3 = Diplont (no H) | ||||||||||||
| Bifurcaria bifurcata | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira tamariscifolia | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira baccata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira nodicaulis | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora grevillei | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora retorta | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Halidrys siliquosa | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum fallax | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum muticum | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis cephalornithos | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis uvifera | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Phyllospora comosa | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Seirococcus axillaris | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Durvillaea potatorum | 12,4 | 2 | 3 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Himanthalia elongata | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Xiphophora chondrophylla | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Hormosira banksii | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Ascophyllum nodosum | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Fucus vesiculosus | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Pelvetia canaliculata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Notheia anomala | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Nemoderma tingitanum | 2,3 | 1 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Cutleria multifida | 5,2 | 1 | 5 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Zanardinia typus | 4,7 | 1 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Tilopteris mertensii | 7,5 | 2 | 8 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 1 | 0 | 1 | 0 |
| Saccorhiza polyschides | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Phyllariopsis brevipes | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | NA | NA | NA | NA |
| Saccorhiza dermatodea | 5,6 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Analipus japonicus | 1,2 | 1 | 7 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Ralfsia fungiformis | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ascoseira mirabilis | 1 | 0 | 8 | 0 | 2 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Agarum clathratum | 7,8 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Laminaria digitata | 2,9 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Alaria spp. | 3 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Undaria pinnatifida | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Ecklonia radiata | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Saccharina latissima | 3,6 | 1 | 9 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Nereocystis luetkeana | NA | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Pelagophycus porra | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Chorda filum | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Hydroclathrus clathratus | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Rosenvingea intricata | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Chnoospora implexa | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | NA | NA | NA | NA |
| Colpomenia peregrina | 1,5 | 1 | 4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Petalonia fascia | 1 | 0 | 7 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Scytosiphon lomentaria | 1,1 | 0 | 8 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Ectocarpus sp. | NA | 1 | 6 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 |
| Petrospongium berkeleyi | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Feldmannia mitchelliae | 1,7 | 1 | 11 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Pylaiella littoralis | 1 | 0 | NA | 0 | 1 | 0 | 1 | 1 | 0 | 1 | NA | NA | NA | NA |
| Elachista fucicola | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Chordaria linearis | NA | 0 | 8 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Dictyosiphon foeniculaceus | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Striaria attenuata | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Asterocladon interjectum | NA | 1 | 7 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Scytothamnus australis | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Splachnidium rugosum | 1,75 | 1 | 6 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Bachelotia antillarum | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Carpomitra costata | 5,5 | 2 | 5 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Perithalia caudata | 2,9 | 2 | 7 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Bellotia eriophorum | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Sporochnus pedunculatus | 7,5 | 2 | 4 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Arthrocladia villosa | 2,1 | 2 | 12 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia menziesii | 2,3 | 2 | Na | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Himantothallus grandifolius | 5 | 2 | 4 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Desmarestia aculeata | 6 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Desmarestia ligulata | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia viridis | 3,9 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Cladostephus spongiosus | 1 | 0 | NA | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Syringoderma phinneyi | 1 | 0 | NA | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Padina spp | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyopteris polypodioides | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyota dichotoma | 13,9 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Ishige okamurae | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Phaeosiphoniella cryophila | 15 | 2 | NA | 1 | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Schizocladia ischiensis | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Vaucheria litorea | 40 | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
H = haploid; D = diploid; outgroup indicated in bold.
List of detailed life cycle and reproductive traits across the brown algal species
| Species | Gamete size ratio (F/M) | Gamete dimorphism | Male gamete size (um) | Isogamy versus anisogamy | Sexual system | HSD or DSD | Co‐sexual versus separate sexes | Type of life cycle | Generation dominance (simple) | Generation dominance | Parthenogenesis capacity | Both parthenogenesis | Parthenogenesis presence/absence | Male parthenogenesis |
| 0 = isogamous | 0 = isogamous | 0 = monoicous | 0 = haploid sex determination | 0 = sexes on same thallus | 0 = H/D with H > D | 0 = haploid‐dominant (or similar dominance) | 0 = H»D | 0 = no parthenogenesis | 0= none or 1 do | 0= no parthenogenesis | 0 = no male parthenogenesis | |||
| 1 = anisogamous | 1 = anisogamous and oogamous | 1 = dioicous | 1 = diploid sex determination | 1 = sexes on separate thalli | 1 = H/D with H = D | 1 = diploid‐dominant | 1 = H=D | 1 = female only | 1 = both do parthenogenesis | 1 = at least female does | 1 = male parthenogenesis | |||
| 2 = oogamous | 2 = monoecious | 2 = H/D with D > H | 2 = D»H | 2 = female and male | ||||||||||
| 3 = dioecious | 3= Diploid | 3 = Diplont (no H) | ||||||||||||
| Bifurcaria bifurcata | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira tamariscifolia | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira baccata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira nodicaulis | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora grevillei | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora retorta | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Halidrys siliquosa | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum fallax | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum muticum | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis cephalornithos | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis uvifera | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Phyllospora comosa | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Seirococcus axillaris | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Durvillaea potatorum | 12,4 | 2 | 3 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Himanthalia elongata | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Xiphophora chondrophylla | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Hormosira banksii | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Ascophyllum nodosum | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Fucus vesiculosus | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Pelvetia canaliculata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Notheia anomala | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Nemoderma tingitanum | 2,3 | 1 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Cutleria multifida | 5,2 | 1 | 5 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Zanardinia typus | 4,7 | 1 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Tilopteris mertensii | 7,5 | 2 | 8 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 1 | 0 | 1 | 0 |
| Saccorhiza polyschides | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Phyllariopsis brevipes | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | NA | NA | NA | NA |
| Saccorhiza dermatodea | 5,6 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Analipus japonicus | 1,2 | 1 | 7 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Ralfsia fungiformis | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ascoseira mirabilis | 1 | 0 | 8 | 0 | 2 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Agarum clathratum | 7,8 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Laminaria digitata | 2,9 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Alaria spp. | 3 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Undaria pinnatifida | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Ecklonia radiata | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Saccharina latissima | 3,6 | 1 | 9 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Nereocystis luetkeana | NA | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Pelagophycus porra | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Chorda filum | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Hydroclathrus clathratus | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Rosenvingea intricata | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Chnoospora implexa | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | NA | NA | NA | NA |
| Colpomenia peregrina | 1,5 | 1 | 4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Petalonia fascia | 1 | 0 | 7 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Scytosiphon lomentaria | 1,1 | 0 | 8 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Ectocarpus sp. | NA | 1 | 6 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 |
| Petrospongium berkeleyi | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Feldmannia mitchelliae | 1,7 | 1 | 11 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Pylaiella littoralis | 1 | 0 | NA | 0 | 1 | 0 | 1 | 1 | 0 | 1 | NA | NA | NA | NA |
| Elachista fucicola | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Chordaria linearis | NA | 0 | 8 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Dictyosiphon foeniculaceus | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Striaria attenuata | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Asterocladon interjectum | NA | 1 | 7 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Scytothamnus australis | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Splachnidium rugosum | 1,75 | 1 | 6 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Bachelotia antillarum | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Carpomitra costata | 5,5 | 2 | 5 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Perithalia caudata | 2,9 | 2 | 7 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Bellotia eriophorum | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Sporochnus pedunculatus | 7,5 | 2 | 4 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Arthrocladia villosa | 2,1 | 2 | 12 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia menziesii | 2,3 | 2 | Na | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Himantothallus grandifolius | 5 | 2 | 4 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Desmarestia aculeata | 6 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Desmarestia ligulata | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia viridis | 3,9 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Cladostephus spongiosus | 1 | 0 | NA | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Syringoderma phinneyi | 1 | 0 | NA | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Padina spp | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyopteris polypodioides | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyota dichotoma | 13,9 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Ishige okamurae | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Phaeosiphoniella cryophila | 15 | 2 | NA | 1 | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Schizocladia ischiensis | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Vaucheria litorea | 40 | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Species | Gamete size ratio (F/M) | Gamete dimorphism | Male gamete size (um) | Isogamy versus anisogamy | Sexual system | HSD or DSD | Co‐sexual versus separate sexes | Type of life cycle | Generation dominance (simple) | Generation dominance | Parthenogenesis capacity | Both parthenogenesis | Parthenogenesis presence/absence | Male parthenogenesis |
| 0 = isogamous | 0 = isogamous | 0 = monoicous | 0 = haploid sex determination | 0 = sexes on same thallus | 0 = H/D with H > D | 0 = haploid‐dominant (or similar dominance) | 0 = H»D | 0 = no parthenogenesis | 0= none or 1 do | 0= no parthenogenesis | 0 = no male parthenogenesis | |||
| 1 = anisogamous | 1 = anisogamous and oogamous | 1 = dioicous | 1 = diploid sex determination | 1 = sexes on separate thalli | 1 = H/D with H = D | 1 = diploid‐dominant | 1 = H=D | 1 = female only | 1 = both do parthenogenesis | 1 = at least female does | 1 = male parthenogenesis | |||
| 2 = oogamous | 2 = monoecious | 2 = H/D with D > H | 2 = D»H | 2 = female and male | ||||||||||
| 3 = dioecious | 3= Diploid | 3 = Diplont (no H) | ||||||||||||
| Bifurcaria bifurcata | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira tamariscifolia | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira baccata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystoseira nodicaulis | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora grevillei | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Cystophora retorta | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Halidrys siliquosa | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum fallax | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Sargassum muticum | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis cephalornithos | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Caulocystis uvifera | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Phyllospora comosa | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Seirococcus axillaris | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Durvillaea potatorum | 12,4 | 2 | 3 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Himanthalia elongata | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Xiphophora chondrophylla | NA | 2 | NA | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Hormosira banksii | NA | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Ascophyllum nodosum | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Fucus vesiculosus | 26 | 2 | 4 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Pelvetia canaliculata | 26 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | NA | NA | NA | NA |
| Notheia anomala | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
| Nemoderma tingitanum | 2,3 | 1 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Cutleria multifida | 5,2 | 1 | 5 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Zanardinia typus | 4,7 | 1 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Tilopteris mertensii | 7,5 | 2 | 8 | 1 | 2 | 1 | 0 | 3 | 1 | 3 | 1 | 0 | 1 | 0 |
| Saccorhiza polyschides | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Phyllariopsis brevipes | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | NA | NA | NA | NA |
| Saccorhiza dermatodea | 5,6 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Analipus japonicus | 1,2 | 1 | 7 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Ralfsia fungiformis | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ascoseira mirabilis | 1 | 0 | 8 | 0 | 2 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Agarum clathratum | 7,8 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Laminaria digitata | 2,9 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Alaria spp. | 3 | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Undaria pinnatifida | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Ecklonia radiata | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Saccharina latissima | 3,6 | 1 | 9 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Nereocystis luetkeana | NA | 2 | 8 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Pelagophycus porra | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Chorda filum | NA | 2 | NA | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Hydroclathrus clathratus | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Rosenvingea intricata | NA | 0 | NA | 0 | NA | NA | NA | 0 | 0 | 0 | NA | NA | NA | NA |
| Chnoospora implexa | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | NA | NA | NA | NA |
| Colpomenia peregrina | 1,5 | 1 | 4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Petalonia fascia | 1 | 0 | 7 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Scytosiphon lomentaria | 1,1 | 0 | 8 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 1 | 1 |
| Ectocarpus sp. | NA | 1 | 6 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 |
| Petrospongium berkeleyi | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Feldmannia mitchelliae | 1,7 | 1 | 11 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| Pylaiella littoralis | 1 | 0 | NA | 0 | 1 | 0 | 1 | 1 | 0 | 1 | NA | NA | NA | NA |
| Elachista fucicola | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Chordaria linearis | NA | 0 | 8 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Dictyosiphon foeniculaceus | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Striaria attenuata | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Asterocladon interjectum | NA | 1 | 7 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Scytothamnus australis | 1 | 0 | 6 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Splachnidium rugosum | 1,75 | 1 | 6 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Bachelotia antillarum | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Carpomitra costata | 5,5 | 2 | 5 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Perithalia caudata | 2,9 | 2 | 7 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Bellotia eriophorum | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Sporochnus pedunculatus | 7,5 | 2 | 4 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Arthrocladia villosa | 2,1 | 2 | 12 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia menziesii | 2,3 | 2 | Na | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Himantothallus grandifolius | 5 | 2 | 4 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| Desmarestia aculeata | 6 | 2 | 5 | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 0 |
| Desmarestia ligulata | NA | 2 | NA | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Desmarestia viridis | 3,9 | 2 | 6 | 1 | 0 | 0 | 0 | 2 | 1 | 2 | NA | NA | NA | NA |
| Cladostephus spongiosus | 1 | 0 | NA | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 1 |
| Syringoderma phinneyi | 1 | 0 | NA | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 |
| Padina spp | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyopteris polypodioides | NA | 2 | NA | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Dictyota dichotoma | 13,9 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Ishige okamurae | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Phaeosiphoniella cryophila | 15 | 2 | NA | 1 | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Schizocladia ischiensis | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| Vaucheria litorea | 40 | 2 | NA | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 0 | 0 |
H = haploid; D = diploid; outgroup indicated in bold.
We coded as ‘isogamous’ algae with physiological and behavioural anisogamy but that have been described as having no size difference between male and female gametes. Note that all brown algae exhibit an asymmetry between male and female, at least at the level of their behaviour, and potentially all the algae scored as isogamous have in fact subtle size differences, but the literature is not detailed enough in this respect. For example, most representatives of the order Ectocarpales have been reported to be ‘isogamous’ (based on observations under the light microscope, but without detailed measurements of gamete size), but some members (Ectocarpus sp., Colpomenia peregrina Sauvageau) have anisogamous male and female gametes (Lipinska et al., 2015). Anisogamy is also present in Asterocladon interjectum Uwai, Nagasato, Motomura et Kogame, which belongs to the last order branching off before the Ectocarpales.
The Laminariales, which is sister group to the clade formed by Ectocarpales and Asterocladales, is almost completely oogamous, with the exception of the genus Saccharina, which has been shown to have eggs with rudimentary flagella (Motomura & Sakai, 1988) being therefore considered strongly anisogamous.
Ancestral state reconstructions and correlation analysis
A likelihood‐based method was used to reconstruct the ancestral state of each of the four life‐history traits. We fitted three different models of trait evolution using the function fitDiscrete from the R package Geiger (Harmon et al., 2008). These models differed in the number of transition rates as follows: equal rates (ER, a single transition rates between all states), symmetric (SYM, forward and reverse transitions are the same) and all‐rates‐different (ARD, each rate is a unique parameter). The corrected Akaike Information Criterion (AICc) was used to compare the alternative models. Each model was estimated on each 100 phylogenetic trees sampled from the posterior distribution to account for uncertainty in tree topology and divergence times. We pruned species from the trees that lacked phenotypic data for the reconstruction of each life‐history trait. State probabilities at the root and transition rates were summarized with the mean and standard deviation values of all iterations, to incorporate phylogenetic uncertainty.
We inferred the number of transitions between states, and their minimum timing, using stochastic character mapping (Huelsenbeck et al., 2003). One hundred stochastic mappings were performed on the posterior sample of trees, and on each, we divided branch lengths into time bins of 1 Myr and recorded the number of transitions from and to each state, in each bin (as described in Serrano‐Serrano et al., 2017). We reported the mean and standard deviation, and the time bin at which 60% of the stochastic mappings had at least one transition event as the onset time for each type of transition.
We assessed correlation between binary life‐history traits using the reversible‐jump MCMC algorithm implemented in BayesTraits V3 (Pagel et al., 2004). This approach compared two models, a null model assuming that the traits had evolved independently and an alternative model assuming that their evolution had been correlated. Each model was run for 10 million generations using the values found in the ancestral state reconstructions for the root state. The two models were compared through their log marginal likelihood by estimating the log Bayes factor. This approach was used to test the correlation between female parthenogenesis and the occurrence of sexes on the same versus separate thalli. Tests showing a significant support for the correlated model were presented as networks of evolutionary transitions using the R package qgraph (Epskamp et al., 2012).
We also used the threshold model of threshBayes in the R package phytools (Revell, 2014), to test for the correlation between a continuous and a discrete variable. The threshold model assumes that the states of discrete phenotypes are governed by an unobserved continuous character called liability. These liabilities are assumed to evolve according to a Brownian motion model (Felsenstein, 2012) and translate into discrete characters once they have passed certain thresholds. We used this model to test the correlation between male gamete size and two discrete traits, male parthenogenesis and sexes on the same or on separate thalli.
For correlation analyses that were significant, we fitted an Ornstein–Uhlenbeck model of evolution by using OUwie from the R package Ouwie (Beaulieu et al., 2012) to further test whether the continuous trait had two discrete selective regimes, determined by the discrete binary trait. We compared the alternative models using the corrected Akaike Information Criterion (AICc)‐selected model.
RESULTS
Ancestral state estimations and transitions between states
Our ancestral state reconstructions inferred equal rates of transition (ER model) between states for all traits, except for the trait ‘sexual system’ where rates were different between states but symmetrical (SYM model gain or loss of a trait). These patterns indicate an overall complex evolutionary history for all sexual traits, involving multiple gains and losses (Figure 2; Table S2).
Life cycle
On the basis of ancestral state reconstructions, the ancestor of all brown algae had a diplohaplontic life cycle, with either isomorphic generations or with a larger and morphologically more complex diploid than haploid generation (Figure 2a; Table S2). Transitions between life cycles occurred most frequently from diploid‐dominant to equally dominant generations, involving a decrease in complexity in terms of the sporophyte morphology (number of different cell types, number of tissues and organs) and a concomitant increase in the complexity of the gametophyte (Figure 2a). A change of dominance from a diploid‐dominant to a haploid‐dominant life cycle occurred for the first time in the last common ancestor of the Scytosiphonaceae family, at least 57.5 (±5.05) My ago, with another independent transition in Cutleria multifida (Turner) Greville (Figure 2a,b). Transitions from a diploid‐dominant to a fully diploid life cycle occurred three times, about 74.5 (±21.41) My ago in the ancestor of the diploid order Fucales, in the ancestor of Ascoseirales and in the ancestor of Tilopteris mertensii (Turner) Kützing. Note however that Tilopteris mertensii is a rather particular case within Tilopteridales (Kuhlenkamp & Müller, 1985), and emergence of monoecy in this species should be interpreted with caution.
Overall, our analysis indicated that the dominance relationship between life cycle generations has been a labile trait in the brown algae, with the diplontic life cycle being the only irreversible state.
Sexual system
The last common ancestor of all brown algae is predicted to have exhibited haploid sex determination and was most likely dioicous (Figure 2a–d, Table S2), but several independent transitions towards monoicy have occurred (Figure 2c, d). The transition from haploid to diploid sex determination, which involved a transition from dioicy to monoecy, occurred independently in the last common ancestor of the order Fucales about 74.5 My ago, in Ascoseirales and in Tilopteris mertensii. The three transitions were simultaneous with the transition from a diplohaplontic to a diplontic life cycle (Figure 2b, d). Dioecy appears to have emerged more recently, around 17.5 Mya, in most families of the order Fucales, with the exception of Sargassaceae and Notheiaceae, which remained monoecious. Further transitions back to monoecy occurred in several genera of the Fucaceae (Xiphophora, Pelvetia and Seirococcus) (Table S2; Figure 2c,d).
Overall, our analysis suggests that the transition to diploid sex determination is irreversible and concomitant with a change in the type of life cycle (from diplohaplontic to diplontic life cycle). In contrast, transitions between separate sexes and combined sexes occurred frequently, either in the haploid or in the diploid phase.
Sexual dimorphism
Regarding gamete size dimorphism, our analysis suggests that oogamy is most likely the ancestral state in the brown algae (Table S2, Figure 2e‐f). The oldest transition took place around 114 My ago, from oogamy to isogamy in the lineage leading to the basal brown algal orders Sphacelariales and Syringodermatales. Another independent transition from oogamy to isogamy took place in the Ascoseirales. Isogamy also emerged in the Ectocarpales, with several independent transitions to anisogamy. Nonetheless, transitions from oogamy to anisogamy were the most frequent transition throughout the phylogeny. Taken together, the results indicate that gamete size dimorphism level is a remarkably labile trait in the brown algae.
Parthenogenesis
The gametes of the ancestral brown algae are predicted to have been unable to perform asexual reproduction through parthenogenesis (Figure 2g‐h). The initial transition from absence of parthenogenesis to female gamete parthenogenesis could not be accurately traced in time along the early diverging branch separating the subclass Fucophycideae from the earlier branching Dictyophycidae. The length of this branch renders identification of the transition during 1 My time bins impossible, as most events fall in different time periods and agreement between reconstructions is very low. The oldest traceable transition that could be timed (85.5 My) and also the one with the highest frequency was from female‐only parthenogenesis to parthenogenesis of both female and male gametes in the order Ectocarpales. A subsequent loss of parthenogenesis can be traced to the last common ancestor of the order Fucales. Note that parthenogenesis is the trait with the lowest sampling, as there are very limited data about this trait in the literature.
Generation dominance and sexual system
Transitions in life cycle phase dominance probabilities were higher in monoicous compared to dioecious species, whatever the direction (q21 and q12 > q43 and q34; Figure 3, Figure S1). In other words, monoicous species exhibit higher turnover rates in terms of generation dominance. Moreover, transitions from monoicous to dioicous states were slightly more frequently observed than transitions from dioicous to monoicous, regardless of life cycle phase dominance (Bayes Factor of 3.51 in favour of the dependent model with q24 ~ q13 > q42 ~ q31, Figure 3).
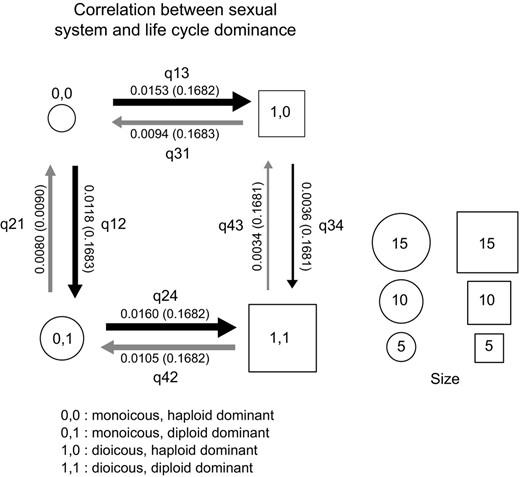
Models of the correlation between traits in brown algae. Binary states are described at the bottom of each panel. Transition rates follow the nomenclature in BayesTraits software (http://www.evolution.rdg.ac.uk/BayesTraitsV3.0.1/BayesTraitsV3.0.1.html). Square boxes indicate the most likely ancestral state (see ancestral state estimations in Methods), sizes of squares/circles represent the abundance of each state in the set of sampled species, and arrow thickness is proportional to the rate of transition between states, which is indicated alongside the arrows (standard deviations in parentheses)
Generation dominance and sexual dimorphism
We tested whether diploid dominance is correlated with an increase in sexual dimorphism. The test of the dependent versus independent model showed that the difference in likelihood was not significant (log BF = −0.1080, Table S3), suggesting that the evolution of these traits is not correlated. Therefore, our data do not support the hypothesis that diploid growth is associated with increased sexual dimorphism. It should be noted, however, that the low number of transitions between both traits may limit the statistical power of this correlation.
Gamete biology and sexual systems
Based on the idea that gamete dimorphism evolved to maximize the chances of gamete encounters, one may hypothesize that separate sexes (dioecy and dioicy) would be associated with small and abundant male gametes, as a mechanism to ensure that the gametes find a partner of the opposite sex when gametes are released into seawater. However, we found no evidence for an association between male gamete size and sexual system (sexes on same vs. different individuals) (Figure S2, Table S3, r = 0.0909).
When gametes are produced by two separate individuals, it may be more difficult for a gamete to find a gamete of the opposite sex than if the same individual produces gametes of both sexes. Accordingly, we hypothesized that parthenogenesis would be favoured in species with separate sexes, as opposed to the situation where male and female gametes are produced by the same individual (note that auto‐incompatibility has not been described in the brown algae, with the exception of one study (Gibson, 1994)). However, we found no evidence that parthenogenesis was more prevalent in species with separate sexes (Table S3).
Finally, we investigated the relationship between the size of male gametes and their parthenogenetic capacity, under the hypothesis that there is a minimum threshold size for male gametes, below which parthenogenesis is not possible. The phylogenetic threshold model indicated that there is a positive correlation between male gamete size and parthenogenetic capacity (Table S3, r = 0.4242); however, the highest posterior density (HPD) interval of this correlation includes zero. We therefore complemented this analysis using an Ornstein–Uhlenbeck (OU) model. The estimated optimal size for nonparthenogenetic male gametes is significantly lower than that of parthenogenetic male gametes (5.49 µm vs. 9.30 µm; Figure S1; Figure 4), further supporting a correlation between male gamete size and male parthenogenesis.
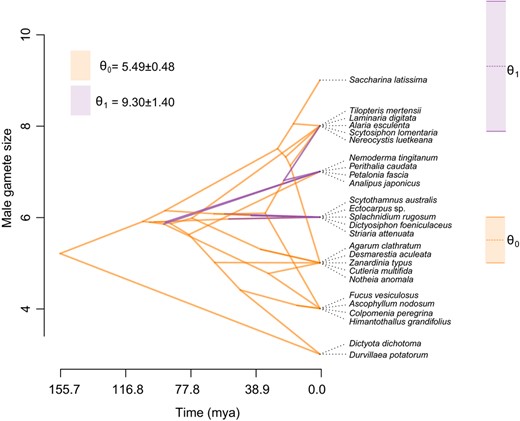
Phenogram for male gamete size and parameter estimation using Ornstein‐Uhlenbeck (OU) model. Colours denote the absence (θ0 in orange) and presence (θ1 in purple) of parthenogenesis of male gametes
DISCUSSION
Several hypotheses have been proposed to explain the evolution of life cycles in eukaryotic lineages (Otto & Gerstein, 2008; Otto & Marks, 1996; Valero et al., 1992). We used 131 species that are representative of the major groups within Phaeophyceae (Silberfeld et al., 2014) to calibrate the divergence time in our phylogeny and used 77 of those species to test some of these hypotheses. The taxonomic sampling in this study greatly exceeds those of previous attempts at understanding the evolution of life cycles in brown algae, with only 14 species (Bell, 1997), as well as recent publications where divergence times were calculated using 44 and 91 species (Kawai et al., 2015; Silberfeld et al., 2014). We found differences in the phylogenetic relationships among brown algae orders compared to Kawai et al., (2015), such as the placement of Scytothamnales and Syringodermatales, or the relative placement of Tilopteridales and Ralfsiales alongside Fucales. These discrepancies could arise from differences in the molecular markers used for the phylogenetic reconstruction, as Kawai et al., (2015) used six chloroplast genes and one mitochondrial gene, whereas we used four chloroplast genes, five mitochondrial genes and one nuclear gene. Future phylogenomic studies will likely shed light on some of these discrepancies. Although Kawai et al., (2015) were able to collect molecular data from Stschapoviales and Onslowiales, which could not be integrated to our dataset, we were able to obtain molecular data from Asterocladales and Nemodermatales, as well as a higher taxonomic sampling from most of the brown algae orders, allowing us to better pinpoint the transition events in trait evolution. We found a limited number of state transitions for each trait throughout the phylogeny, which limited the statistical power of our correlation analyses, and thus the extent of our conclusions. Nonetheless, we were able to find some instances of correlated evolution between traits that helped us take a step forward in understanding some aspects of the evolution of life cycles.
We found several coincidental trait transitions leading to the common ancestor of Fucales ~74.5 My ago, after its split with Nemodermatales. During this time, sea levels rose 100 m above present‐day levels (Surlyk & Sørensen, 2010). The extant species of Fucales inhabit the intertidal and subtidal zones (Schiel & Foster, 2006), so the rising of sea levels possibly opened new ecological niches for these taxa, allowing the emergence of evolutionary novelty. We found another important transition at least ~57.5 My ago from a diploid‐dominant to a haploid‐dominant life cycle in the last common ancestor of Scytosiphonaceae. This event coincides with the Paleocene–Eocene Thermal Maximum, a major geological event where temperature increased and large amounts of carbon were introduced to the oceans through volcanic activity, leading to many changes in marine ecosystems (Ma et al., 2014).
Is diploid growth indirectly associated with sexual reproduction as a way to complement deleterious mutations?
Our results indicate that the ancestral brown algae likely had a diplohaplontic life cycle with similar diploid and haploid dominance (i.e. similar size and complexity of the gametophyte and sporophyte generations). Over evolutionary time, the diploid phase became dominant in some clades, whereas other clades evolved towards greater haploid dominance. Several theories have been proposed to explain evolution towards either a dominant haploid or a dominant diploid phase in the life cycle (Otto & Gerstein, 2008). Hypotheses based on the effect of deleterious alleles have proposed that being diploid generally increases mean fitness due to the masking of deleterious alleles (due to complementation of these alleles by nonmutant alleles), whereas developing as a haploid allows more efficient purging of deleterious alleles because they are exposed to selection (Otto & Goldstein, 1992; Rescan et al., 2016; Scott & Rescan, 2017). The balance between these two forces determines whether evolution proceeds towards an increase of the haploid or the diploid phase and depends critically on the importance of sexual exchanges within populations. Indeed, under higher rates of inbreeding or asexual reproduction, the benefit of purging deleterious alleles remains associated with alleles increasing the haploid phase; therefore, haploidy is favoured. In contrast, outcrossing and/or more frequent sex tend to favour diploidy (Otto & Marks, 1996). In accordance with this hypothesis, our results show that the last common ancestor of Fucales transitioned to a loss of parthenogenesis alongside a fully diplontic life cycle, whereas the haplontic family Scytosiphonaceae conserved both female and male parthenogenesis. Nonetheless, a direct correlation between deleterious mutations and phase dominance remains to be tested. To test this hypothesis, future studies should look for correlations between the dominance of the haploid or diploid phase with changes in the accumulation of nonsynonymous mutations in the nuclear genome, as diploid‐dominant taxa are expected to maintain a higher fitness than haploid‐dominant taxa after the accumulation of substitution events (Rescan et al., 2016).
Very few estimates of inbreeding coefficients or rates of asexual reproduction are available for brown algae, with most of them being done in Laminariales and Fucales (Bringloe et al., 2020). However, this idea was tested by Bell (1997) by looking at the correlation between the sexual system of a species (monoicous or dioicous) and the relative dominance of the haploid and diploid phases of the life cycle, assuming that monoicous species will tend to be more inbred due to selfing. At the time, Bell concluded that monoicous species did not tend to have more dominant haploid phases (Bell, 1997). In contrast to Bell's analysis, which was based on a small number of brown algal species, our results do appear to support Otto and Marks' (1996) ideas, at least to some extent, because transitions towards dominance of the haploid phase were found to be more frequent when the sexual system was monoicous, consistent with the idea that monoicy is correlated with haploid growth. Generating novel data on estimates of inbreeding coefficients within natural populations of monoicous species would be extremely valuable to shed further light into these phenomena.
Somatic mutations have been proposed as another possible source of selection for diploidy, as these mutations should have a lower impact on the fitness of diploid organisms (Otto & Gerstein, 2008). This idea is consistent with the general observation that larger organisms tend to be diploid rather than haploid, as in the case of vascular against nonvascular plants (Schoen & Schultz, 2019). Indeed, this pattern also holds true for the brown algae, since all the largest brown algae (e.g., Laminariales, Fucales) have a dominant diploid phase. Empirical estimates of somatic mutations in haplontic and diplontic taxa and their effects on fitness could help test this hypothesis (Yu et al., 2020).
Is diploid growth associated with increased gamete dimorphism?
The theory for the evolution of gamete dimorphism based on the trade‐off between gamete number and offspring fitness predicts that dimorphism may evolve when zygote size has a strong effect on fitness (i.e. when offspring fitness increases more than linearly in relation to zygote size). Accordingly, one may predict that if a larger zygote size is needed for larger diploid development, increased diploid growth would favour higher levels of gamete dimorphism (Parker et al., 1972). Bell (1994) proposed an alternative theory that also predicts a correlation between diploid growth and gamete dimorphism, in which the direction of causality is reversed (i.e. sexual selection caused by gamete dimorphism favouring diplontic cycles in order to increase genetic differences between gametes produced by the same organism). Our results show that diplontic brown algal species are mostly oogamous, suggesting a link between strong sexual dimorphism and diploidy. However, those associations may not reflect a general tendency, because they are mainly based on the Fucales and Tilopteris mertensii, whereas Ascoseirales transitioned towards isogamy alongside a diplontic life cycle. Therefore, more analyses are needed to explore the idea that diploid growth is associated with increased sexual dimorphism.
Evolution of sexual systems in the brown algae
Our results indicate that the ancestral sexual system of brown algae corresponds to haploid sex determination and dioicy, with several transitions towards monoicy having occurred independently over evolutionary time. Transition towards a diplontic life cycle in the Fucales, Ascoseirales and Tilopteris mertensii appears to have involved a monoecious/hermaphrodite intermediate state, with subsequent independent re‐emergence of dioecy in some lineages of Fucales. It is interesting to note that transitions from separate sexes to co‐sexuality are relatively frequent in haploid sexual systems, which contrasts with what is the most commonly accepted direction of evolution in clades with diploid sex determination (i.e. monoecy to dioecy). Note, however, that although dioecy was considered to be an evolutionary dead end in angiosperms (Vamosi & Otto, 2002), more recent phylogenetic analyses are challenging this conclusion, and the idea that reversals to monoecy in angiosperms may be more frequent than thought before is increasingly becoming accepted (Käfer et al., 2017; Pannell, 2017).
In diploid sexual systems, two main selective effects have been proposed to explain transitions from co‐sexuality to dioecy: inbreeding depression (selfing is less likely to occur when male and female gametes are produced by separate individuals) and the effect of trade‐offs between male and female fitness (Charlesworth & Charlesworth, 1978; Charnov, 1982). In haploid sexual systems, the opposite transition (from separate sexes towards co‐sexuality) could also be caused in principle by a change in the shape of the trade‐off between male and female reproductive success (leading to a higher fitness of gametophytes producing both types of gametes) or by selection for inbreeding (selfing), either through the automatic transmission advantage associated with selfing (Fisher, 1941), or for reproductive assurance when population density is low. Note, however, that parthenogenesis occurs in all monoicous species, and this process may represent an alternative way of dealing with mate limitation and reproductive assurance. Assuming that selfing occurs following transitions to monoicy, such transitions should occur more easily when inbreeding depression is low. More transitions to monoicy in taxa with a prolonged haploid phase (if at least a proportion of the deleterious alleles affecting the fitness of diploids will be purged during the haploid phase of the life cycle) would therefore be expected, but this is not what we observe in our results (q31 < q42, Figure 3). Examining the proximate mechanisms involved in the transitions between separate sexes and co‐sexuality in both haploid and diploid systems and more natural population data, for example in populations with different densities, would be valuable to shed light on the mechanisms and evolutionary forces driving the shifts among sexual systems in the brown algae.
Anisogamy is ancestral in the brown algae
In agreement with the tendencies observed by Silberfeld et al., (2010), our analysis points towards an oogamous ancestor of brown algae, with several independent transitions towards anisogamous and isogamous clades. This stands in contrast to theoretical scenarios representing the evolution of gamete dimorphism from an isogamous ancestor (Lehtonen & Kokko, 2011; Parker et al., 1972; Randerson & Hurst, 2001), in which isogamy is ancestral and anisogamy represents an intermediate step during the process of increased gametic differentiation. The transition from oogamy to isogamy has also been reported in diatoms, where possible explanations included reproductive adaptations such as physical proximity, mucilage envelopes or copulation tubes that facilitate the success rates in the pairing of gametes, changing the selective pressures that favour oogamy (Edlund & Stoermer, 1997). Theories based on disruptive selection caused by a trade‐off between the number of gametes produced and zygote size (Bulmer & Parker, 2002; Parker et al., 1972) have shown that the shape of the relation between zygote size and fitness is critical for the evolution of gamete dimorphism. It would be interesting to explore whether a change in the relation between zygote size and fitness (e.g. due to a decrease in size of the diploid organism) may favour transitions from oogamy to anisogamy or isogamy. This type of evolutionary mechanism may generate a positive correlation between the degree of gamete dimorphism and the relative importance of the diploid phase, leading to an inversion of the causal relationship in Bell's (1994, 1997) hypothesis mentioned above, that is decrease in the size of the diploid organism would drive a decrease in gamete dimorphism.
The evolution of anisogamy requires some level of gametic competition and limitation (Lehtonen & Kokko, 2011). Therefore, it is likely that in specific conditions the system may return to isogamy or near isogamy, for instance if there is a low level of gamete competition or if there is no gamete limitation.
Evolution of gamete size and parthenogenetic capacity
Our results point towards a possible correlation between the capacity of brown algae to perform male parthenogenesis and male gamete size. There are marked differences between the relative parthenogenetic capacities of male and female gametes in isogamous, anisogamous and oogamous brown algal species (Luthringer et al., 2014). It has been suggested that increased gamete size leads to increased parthenogenetic capacity, up to a point, but that in oogamous species, the large female gamete loses its flagella becoming specialized for zygote production, often losing its capacity to initiate parthenogenetic development (Luthringer et al., 2014).
Our results indicate that the gametes of the common ancestor of brown algae were most likely unable to perform asexual reproduction through parthenogenesis, suggesting that the emergence of gamete parthenogenetic capacity was derived, perhaps as an adaptive trait in situations where mates are limited, such as in marginal populations (Bierzychudek, 1985; Oppliger et al., 2014). Data from field populations of a range of species would be needed to further understand whether parthenogenesis is adaptive. It is noteworthy that parthenogenetic capacity is assessed under laboratory conditions and that the contribution of parthenogenesis to recruitment in natural populations would be worth exploring further (Oppliger et al., 2007, 2014). A recent study in field populations of Ectocarpus showed no evidence that parthenogenesis plays a significant role under field conditions (Couceiro et al., 2015). In contrast, studies in field populations of another brown algal species Scytosiphon lomentaria (Lyngbye) J. Agardh suggested that parthenogenesis is prevalent in field populations (Hoshino et al. 2019). Interestingly, female‐only parthenogenetic populations have larger gamete sizes relative to ‘sexual’ populations of the same species, consistent with a link between gamete size and parthenogenetic capacity, and opening the possibility that parthenogenesis may be an adaptive trait.
Overall, our results reveal that the emergence and dominance of certain life‐history traits within the brown algae are associated throughout the evolutionary history of this lineage. These correlations uncovered clues about the evolutionary processes governing the origin of fundamental biological features in eukaryotes, such as the dominance of the haploid or diploid phases throughout the life cycle, the emergence and loss of sexual dimorphism and the evolvability of parthenogenesis. Moreover, our reconstruction of ancestral states highlights the unusual evolutionary history of brown algae, showing that oogamous lineages can revert to an isogamous state, that parthenogenesis can emerge in male gametes and that transitions between monoicous and dioicous states can be very dynamic, with dioicous transitions being more frequent. Despite the considerable rate of trait transitions within the phylogeny, we found that diplontic life cycles and diploid sex determination are both stable and seemingly irreversible, suggesting they are highly advantageous. The advantage of complementing deleterious mutations in diplontic taxa (Otto & Goldstein, 1992; Rescan et al., 2016; Scott & Rescan, 2017) and its concomitance with diploid sex determination may be responsible for the irreversibility of these traits and would likely explain why they are the prevailing biological traits in the plant and animal lineages.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR'S CONTRIBUTIONS
SH, RL, AFP, CD, DR, SMC performed the bibliographic searches; SH, MSS, NS, JBR performed the phylogenetic analysis; SH, MSS, MV, NS, SMC, DR, JMC, CD analysed data; SMC, DR, MV, JMC, NS conceived the study; SMC coordinated the study; SH, MSS, JBR drafted and SMC wrote the final version of the manuscript, which all authors critically revised.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/jeb.13880.
DATA AVAILABILITY STATEMENT
The nucleotide sequences used for the phylogenetic reconstruction are available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession numbers published by Silberfeld et al., (2010, 2014) and in Table S1. The concatenated multiple sequence alignment and the Common Ancestor Tree obtained from BEAST are available in TreeBASE (submission ID S28254).
REFERENCES
Author notes
Joint first authors.
Funding information
This work was supported by the CNRS, Sorbonne Université and the ERC (grant agreement 638240)
Supplementary data
Supplementary Material
Dataset S1




