Abstract
The Gram-negative bacteria Yersinia pestis, causative agent of plague, is extremely virulent. One mechanism contributing to Y. pestis virulence is the presence of a type-three secretion system, which injects effector proteins, Yops, directly into immune cells of the infected host. One of these Yop proteins, YopJ, is proapoptotic and inhibits mammalian NF-κB and MAP-kinase signal transduction pathways. Although the molecular mechanism remained elusive for some time, recent work has shown that YopJ acts as a serine/threonine acetyl-transferase targeting MAP2 kinases. Using Drosophila as a model system, we find that YopJ inhibits one innate immune NF-κB signaling pathway (IMD) but not the other (Toll). In fact, we show YopJ mediated serine/threonine acetylation and inhibition of dTAK1, the critical MAP3 kinase in the IMD pathway. Acetylation of critical serine/threonine residues in the activation loop of Drosophila TAK1 blocks phosphorylation of the protein and subsequent kinase activation. In addition, studies in mammalian cells show similar modification and inhibition of hTAK1. These data present evidence that TAK1 is a target for YopJ-mediated inhibition.
Keywords: Drosophila immunity, innate immunity
YopJ is one of six effector proteins (Yops) injected into the host cell cytoplasm during a Yersinia pestis infection. YopJ has been shown to be both proapoptotic and to inhibit proinflammatory signal transduction (1–5). However, the precise targets and mechanism used by YopJ to interfere with these signaling pathways have been controversial. Initially YopJ was proposed to act as an ubiquitin-like protein protease, cleaving the ubiquitin-like protein SUMO from unidentified conjugated targets (6). Subsequently, it was argued that YopJ acted as a deubiquitinase, removing critical polyubiquitin chains from the essential NF-κB/innate immune signaling pathway protein TRAF6 (7, 8). Recent biochemical evidence from two groups strongly argues that YopJ has a completely novel and unpredicted function, that of a serine/threonine acetyltransferase. In this role, YopJ acetylates serines and threonines on various mitogen-activated protein kinase kinases (MAP2Ks). This acetylation blocks the phosphorylation and activation of these kinases, thus neutralizing these signaling pathways (9, 10). Interestingly, the Yersinia enterocolitica homolog, YopP, has been shown to inactivate the mammalian MAP3 kinase, hTAK1; however, the mechanism of suppression is not yet established (11, 12).
Unlike mammals, Drosophila lack a fully developed adaptive immune system and instead rely largely on two innate immune signaling pathways, the Toll and IMD pathways, to control antimicrobial peptide gene expression and other defense responses. Infection by many Gram-positive bacteria (with Lysine-type peptidoglycan) or by fungi leads to the proteolytic cleavage of pro-Spätzle. Once cleaved, active Spätzle then binds the receptor Toll, which initiates a signaling cascade through the adaptor proteins MyD88 and Tube to the kinase Pelle (13, 14). As a consequence, the Drosophila IκB homolog Cactus is phosphorylated, ubiquitinated, and degraded by the proteosome, which allows the NF-κB proteins DIF and Dorsal to translocate into the nucleus and activate antimicrobial peptide gene synthesis. Conversely, the IMD immune signaling pathway is activated by DAP-type peptidoglycan (PGN), common to Gram-negative bacteria and certain Gram positives (15). DAP-type PGN is recognized by two receptors, PGRP-LC and PGRP-LE, which signal the caspase-8–like DREDD to cleave imd protein. Cleaved-IMD is then K63-polyubiquitinated through its association with the E3 ligase DIAP2 (16). These K63-polyubiquitin chains are proposed to then recruit and activate the downstream MAP3 kinase dTAK1, as reported for mammalian NF-κB signaling pathways (17, 18). Once activated, dTAK1 initiates two downstream arms of the IMD pathway. In the Relish/NF-κB arm, dTAK1 phosphorylates and activates the IKK complex leading to the subsequent phosphorylation and activation of the NF-κB protein Relish, which drives the expression of a battery of antimicrobial peptide genes (19, 20). In the second arm of the IMD pathway, dTAK1 also activates the JNK kinase Hemipterous, which, in turn, phosphorylates the JNK homolog Basket, leading to the activation of AP1 transcription factors and induction of various immune genes (19, 21).
Both the Drosophila IMD and Toll pathways show homology to mammalian innate immune signaling pathways, and either could be targets of YopJ-mediated inhibition. However, we find that only the IMD pathway is sensitive to YopJ and that the presence of YopJ results in the serine/threonine acetylation of dTAK1 and subsequent inhibition of this kinase. This inhibition is sufficient to block downstream signaling to both the JNK and Relish/NF-κB branches of the IMD pathway. Furthermore, we demonstrate similar YopJ-mediated inhibition of hTAK1 in mammalian cells. These data demonstrate that the MAP3 kinase TAK1 is a target of YopJ-mediated inhibition and suggests that the broad innate immune inhibitory activity of YopJ results from its ability to inactivate this MAP3K.
Results
YopJ Blocks the IMD Innate Immune Signaling Pathway.
To develop Drosophila as a model system for the study of YopJ-mediated innate immune inhibition, S2* stable cell lines inducibly expressing either wild-type YopJ (YopJWT) or a catalytically inactive mutant (C172A, YopJCA) under the control of the metallothionein promoter were generated. In these cells, the addition of copper sulfate induced YopJ expression, whereas in the absence of copper, no protein was detected (Fig. 1A, Lower). These cells were stimulated with Spätzle-C106 (22) or DAP-type PGN to activate the Toll or IMD pathways, respectively. Toll signaling was monitored by Northern blotting for Drosomycin message, whereas IMD signaling was monitored by probing for Diptericin (and, to a lesser degree, Drosomycin) (Fig. 1A). YopJ had no effect on the Spätzle-induced expression of Drosomycin, whereas PGN-induced Diptericin and Drosomycin expression was dramatically inhibited. The failure to induce Diptericin was clearly linked to the presence of YopJWT, because YopJCA was not inhibitory and without copper pretreatment Diptericin induction was robust. Together, these results demonstrate that YopJ is a potent inhibitor of IMD, but not Toll, signaling in Drosophila cells.
Fig. 1.
YopJ inhibits Drosophila IMD but not Toll immune signaling and functions between IMD and JNK. (A) S2* cells stably expressing YopJWT or YopJCA under control of the metallothionein promoter were pretreated with copper (to activate expression of YopJ) or not, before stimulation with Spätzle (Left) or DAP-type PGN (Right). Activation of immune signaling was monitored by Northern blotting for Diptericin and Drosomycin RNA. (B) Expression of YopJWT or YopJCA was induced with the addition of copper sulfate in S2* cells before stimulation with PGN. Ubiquitination of IMD was monitored by IMD immunoprecipitation followed by immunoblotting for ubiquitin (Upper). Anti-IMD blotting was used as a loading control and also to examine PGN-induced IMD cleavage. Anti-FLAG probing was used to verify the presence/absence of FLAG-YopJ.  marks unmodified full-length IMD,
marks unmodified full-length IMD,  highlights phosphorylated IMD, and
highlights phosphorylated IMD, and  marks the cleaved-IMD products. (C) Phospho-JNK was monitored in whole-cell lysates. Full-length JNK blot serves as a loading control. (D) IKK activity was monitored in S2* cells expressing YopJWT or YopJCA after stimulation with PGN. IKKγ blot serves as an immunoprecipitation control.
marks the cleaved-IMD products. (C) Phospho-JNK was monitored in whole-cell lysates. Full-length JNK blot serves as a loading control. (D) IKK activity was monitored in S2* cells expressing YopJWT or YopJCA after stimulation with PGN. IKKγ blot serves as an immunoprecipitation control.
For studies in whole animals, we generated YopJ transgenic flies. Using the dual GMR/UAS promoter system (pGUS; ref. 23) (Fig. S1A), we generated flies constitutively expressing wild-type or mutant YopJ in the eye, and at any other location/time with appropriate Gal4 “drivers.” To monitor the effect of YopJ on the humoral systemic immune response, the fat body-specific yolk-Gal4 driver was used. After infection with live Escherichia coli, control animals (female yolk-gal4 driver flies or YopJWT flies containing no Gal4 driver) showed robust IMD pathway activation, as assayed by Northern blotting for Diptericin. However, the yolk-gal4;UAS-YopJWT females exhibited markedly reduced Diptericin expression (Fig. S1B), consistent with our cell culture data. Together these observations show that YopJ blocks IMD signaling in both tissue culture and whole animals.
Interestingly, moderate expression of YopJWT in the developing eye imaginal disk (from the GMR element in the pGUS transgene) led to a rough and reduced eye phenotype, which was not observed in YopJCA flies (Fig. S1C). Multiple transgenic insertion lines were generated for both wild-type and mutant YopJ. Although the penetrance of the rough eye phenotype varied among the YopJWT lines (as shown in Fig. 1C, columns 1 and 2), no rough/reduced eye phenotype was present in any lines expressing YopJCA. The rough and reduced eyes found in YopJWT flies were further enhanced by driving higher levels of YopJWT via an additional GMR-GAL4 driver (Fig. S1C, Lower). Unlike the Toll pathway, the IMD pathway is not involved in development and most IMD pathway mutants are viable to adulthood, with normal eyes. Therefore, it seems likely that the developmental defects seen in YopJWT flies are a consequence of inhibition of other MAP kinase pathways important for eye development. Further work will be required to identify these kinases.
Having established that YopJWT inhibits the IMD signaling pathway, we next sought to determine which component(s) of this signaling pathway are targeted. We have shown that the cleavage and subsequent ubiquitination of the imd protein are crucial events during signaling (16). Because YopJ was proposed to act as a ubiquitin-protein protease (7, 8), we examined whether either of these IMD modifications were altered in stable cell lines expressing YopJ. After stimulation with PGN, comparable levels of IMD cleavage and ubiquitination were detected in both the YopJWT and YopJCA expressing cells, in the presence or absence of copper, indicating that YopJ does not function as an IMD-specific ubiquitin-protein protease (Fig. 1B).
However, downstream signaling events in both the JNK and NF-κB arms of the IMD pathway were clearly inhibited by YopJ. For example, the Drosophila JNK pathway, as assayed by immunoblotting for PGN-induced phospho-JNK, was blocked by YopJWT (Fig. 1C). In the NF-κB/Relish branch, activation of the Drosophila IKK complex was also inhibited by YopJWT, but not by YopJCA (Fig. 1D). Together with the inhibition of AMP gene induction, these data indicate that YopJ acts by inhibiting the IMD pathway downstream of IMD cleavage and ubiquitination but upstream or at the level of JNK phosphorylation and IKK activation. Although Orth and colleagues have argued that both NF-κB and JNK signaling are inhibited by YopJ-mediated acetylation of multiple MAP2 kinases (10), an alternate possibility is that YopJ also inhibits a common upstream protein that is required for both NF-κB and JNK activation, such as dTAK1.
YopJ Inhibits dTAK1 Activation.
To examine whether YopJ may be directly inhibiting the kinase dTAK1, we next coexpressed both dTAK1 and YopJ (WT or CA) in double-stable S2* cells. In these cells, dTAK1 and IKK kinase activities were monitored with IP-kinase assays by using recombinant MKK6K82A (a mammalian TAK1 target) or recombinant Relish as substrates, respectively. In S2* cell lines, overexpression of dTAK1 was sufficient to activate both dTAK1 and the downstream IKK complex (Fig. 2A, rows 1 and 4). When both dTAK1 and YopJWT were coexpressed, the activity of both kinases was inhibited, whereas coexpression of YopJCA resulted in no discernible reduction in signal. Interestingly, dTAK1, expressed alone or in combination with YopJCA, migrated as a tight doublet at a molecular mass just below 105 kDa, whereas in concert with YopJWT, dTAK1 migrated noticeably faster, closer to 85 kDa (Fig. 2A, row 2). When optimized for better resolution, immunoblot analysis showed that overexpressed dTAK1 alone, or in the presence of YopJCA, migrates as a doublet at ∼95 and 105 kDa. In the presence of YopJWT, dTAK1 runs faster at a molecular mass of ∼85 kDa (Fig. 2A, row 3). The predicted molecular mass for Drosophila TAK1 is 76 kDa. Together, these data strongly indicate that YopJ directly interferes with the activity of dTAK1 and that this interference is likely the result of posttranslational modification(s).
Fig. 2.
YopJ inhibits Drosophila TAK1 kinase activity. (A) dTAK1 was overexpressed in S2* cells alone or in the presence of YopJWT or YopJCA, and activity was monitored by IP-kinase assays with catalytically inactive MKK6 serving as a substrate (row 1). As a control, immunoprecipitated FLAG-TAK1 were immunoblotted with anti-FLAG to verify kinase capture and banding pattern (row 2). To more clearly observe YopJ-mediated alteration in the dTAK1 banding pattern, FLAG-TAK1 was immunobloted directly from lysates under optimized conditions (row 3). IKK activity was similarly monitored by immunoprecipitation using an endogenous IKKγ antisera and the substrate Relish (row 4). Immuoprecipitated IKKγ samples were immunoblotted with IKKγ antisera to verify capture (row 5). The presense of FLAG-YopJ was also monitored by FLAG IP/immunoblot (row 6). (B) Anti-FLAG (TAK1) immunoblot of lysates from S2* cells expressing FLAG-TAK1 alone or in the presence of YopJWT or YopJCA were treated (or not) with λ-phosphatase. (C) Activation of endogenous dTAK1 was monitored by IP-kinase assays, with rMKK6 as substrate, from S2* cells expressing either YopJWT or YopJCA. Anti-FLAG immunoblot serves as a control for the presence of YopJ.
To determine whether the observed Drosophila TAK1 bands represent phosphorylated forms, we treated lysates with λ-protein phosphatase. Phosphatase treatment resolved the doublets observed in the dTAK1 alone and dTAK1 with YopJCA samples from ∼95/105 kDa to ∼85/93 kDa. In the presence of YopJWT, no change in the migration of dTAK1 was detected with phosphatase treatment (Fig. 2B). These data show that active dTAK1, (when expressed alone or with the inactive YopJCA) is phosphorylated; however in the presence of YopJWT, dTAK1 is inactive and unphosphorylated.
To analyze the effect of YopJ on endogenous dTAK1, IP-kinase assays were undertaken with an anti-dTAK1 antibody (16). Cells expressing YopJWT showed little PGN-induced TAK1 kinase activity, whereas cells similarly expressing YopJCA showed activity comparable to parental S2* cells (Fig. 2C). Although our anti-dTAK1 antibody is effective for immunopreciptiation, as monitored by endogenous dTAK1 IP-kinase assays, it is not very useful for immunoblotting (16). Therefore, we used an RNAi approach to validate this assay. RNAi treatment targeting dTAK1 was able to inhibit endogenous dTAK1 kinase activity in this IP-kinase assay and accumulation of phospho-JNK after immune induction (Fig. S2). Together, the results with both endogenous or overexpressed dTAK1 strongly suggest that YopJ interferes with this MAP3 kinase.
YopJ Acetylates dTAK1.
An alignment of Drosophila and mammalian TAK1 shows that three phospho-acceptor sites found in the activation loop of mammalian TAK1 (T184, T187, and S192) and most of the surrounding residues are highly conserved (Fig. 3A). In mammals, alanine substitution of any of these serine or threonine residues is sufficient to ablate kinase activity and block downstream signaling (24–28). To identify the phosphorylated residues of Drosophila TAK1 and map the molecular changes induced by YopJ, FLAG-TAK1 was isolated from lysates prepared from cells expressing dTAK1 alone or in combination with YopJWT or YopJCA. These samples were then subjected to microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (MS/MS).
Fig. 3.
YopJ acetylates Drosophila TAK1. (A) Alignment of human and Drosophila TAK1 activation loops. The established phosphorylation sites on mammalian TAK1 are indicated (P). (B) Summary of tandem MS identification of Drosophila TAK1 activation loop. Phosphorylated (P) and acetylated (Ac) residues are indicated. All phospho-peptide and acetyl-peptide residues can be found in Tables S1–S3. In both A and B, conserved residues are shaded in gray. (C) Activity of dTAK1 alanine substitutions in the presence or absence of YopJ assayed by IP/cold kinase assay (row 1). Immunoblots for FLAG-TAK1 and FLAG-YopJ verify the presence of the respective proteins (rows 2 and 3). (D) Activity of dTAK1 glutamic acid substitutions by IP/cold kinase assay [IP/KA-followed by immunoblot for phospho-MKK6 (S207)] (row 1). FLAG immunoblot verifies the expression of dTAK1 protein (row 2).
Within the activation loop of Drosophila TAK1, a single phosphorylation, at S176 (which aligns to S192 of the mammalian protein), was identified by MS/MS (Fig. 3B). A number of other phosphorylation sites were found outside the activation loop (see Table S1 for the full list of phosphoresidues). Interestingly, no phosphorylation was detected in the activation loop of dTAK1 when coexpressed with YopJWT, and overall phosphorylation was greatly reduced. When coexpressed with YopJCA, dTAK1 was phosphorylated on a number of residues, including all of the sites identified when TAK1 was expressed alone and some additional sites outside the activation loop (Table S1). The relevance of these additional phosphorylation sites is unclear; however, it is important to note that when expressed with YopJCA, dTAK1 remains fully functional. Given that YopJ is thought to act as a serine/threonine acetyl-transferase (9, 10), mass spectrometry was also used to detect acetylation. In the presence of YopJWT, dTAK1 showed clear acetylation of both T168 and T171 and possible acetylation of S167 and S176, all within the activation loop and at residues highly conserved with mammalian TAK1 (Fig. 3B). However, dTAK1 alone or in the presence YopJCA displayed no detectible acetylation (see Table S2 for a full list of acetyl residues). Together, these data suggest that acetylation of dTAK1 threonines T168 and T171 (and possibly S167 and S176) by YopJ inactivates the kinase, which leaves it in an unphosphorylated state.
Previous studies with mammalian TAK1 indicate that the residues equivalent to Drosophila TAK1 T168, T171, and S176 (in mammalian TAK1: T184, T187, and S192) play important roles in the kinase activity of hTAK1 (24–28). To elucidate the role that these residues play in Drosophila TAK1 activity, we generated various substitution mutations at these sites. As shown previously, overexpression of wild-type dTAK1 is sufficient to activate dTAK1 kinase activity, as monitored by cold-IP-kinase assay (e.g., immunoblotting with anti-pMKK6; Fig. 3C). Interestingly, substitution of T168 to alanine did not affect kinase activity, and this mutant is still subject to YopJ-mediated inhibition, likely because T171 is still a target for acetylation and, as such, is sufficient for inhibition of the kinase. Conversely, substituting either T171 or S176 to alanine renders dTAK1 inactive (Fig. 3C), indicating that these residues are necessary for proper dTAK1 function. These data are consistent with studies of mammalian TAK1 that indicate the corresponding residues, T187 and S192, are essential for activation of the kinase, whereas T184 plays a minor role (24–28). In an attempt to bypass YopJ-mediated acetylation of these residues, we also generated a second set of Drosophila TAK1 mutants in which these serine and threonines were changed to a phosphomemetic acidic residue. Unfortunately, substitution of T168, T171, or S176 to glutamic acid rendered TAK1 inactive (Fig. 3D). All together, this analysis of dTAK1 substitution mutants indicates that residues T171 and S176 are essential for kinase activity, and that substitution or acetylation of these residues impairs the function of dTAK1.
YopJ Inhibits and Acetylates Mammalian TAK1.
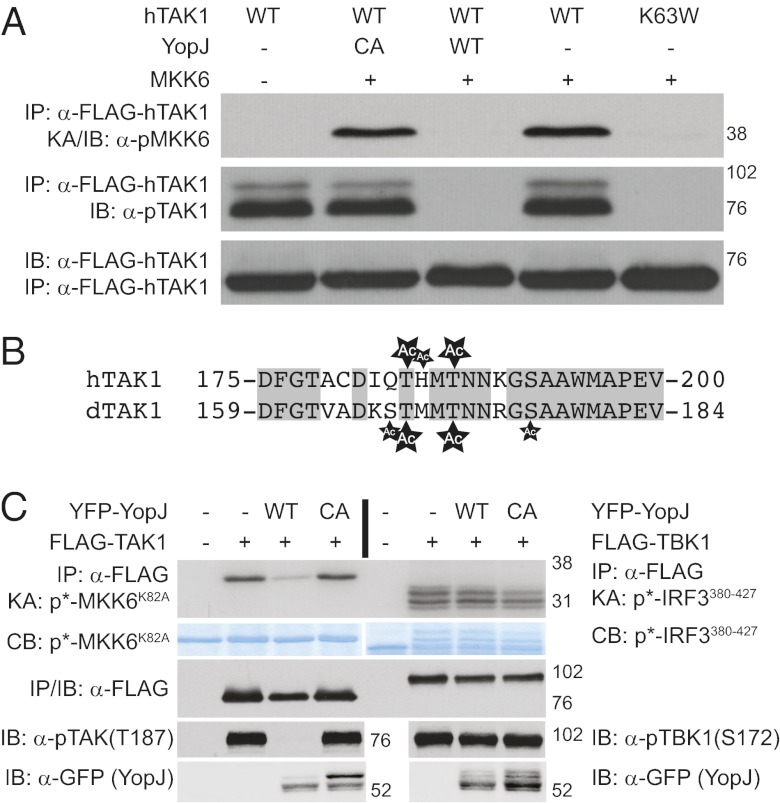
With the discovery that YopJ inactivates Drosophila TAK1, we sought to determine whether YopJ had a similar effect on mammalian TAK1. To that end, transient expression of wild-type mammalian TAK1 in human 293T cells resulted in kinase activation and autophosphorylation, as monitored by cold-kinase assay using MKK6 substrate or immunoblotting for phospho-hTAK1, respectively (Fig. 4A). Interestingly, when wild-type YopJ was coexpressed with hTAK1, the kinase failed to activate, similar to the effects observed with YopJ and Drosophila TAK1. As controls, an inactive hTAK1 mutant (TAKK63W) also failed to autophosphorylate or activate, whereas the coexpression of YopJCA had no effect.
Fig. 4.
YopJ inhibits and acetylates mammalian kinases. (A) Wild-type or K63W hTAK1 was transiently expressed in human HEK 293T cells in the presence or absence of YFP-tagged YopJ. hTAK1 activity was monitored by cold IP-kinase assay (row 1). Phospho- and total hTAK1 were also monitored by IP/immunoblot (rows 2 and 3). (B) Alignment of hTAK1 and dTAK1 activation loops. Conserved residues are boxed (gray). Acetylated residues, as detected by MS/MS, are marked (Ac). Small Ac indicates residues that showed ambiguous acetylation. (C) hTAK1 (Left) or TBK1 (Right) were expressed in human HEK 293T cells in the presence or absence of wild-type (WT) or inactive (CA) YopJ. Kinase activities of hTAK1 and TBK1 were assayed by IP kinase assay and with phosphospecific hTAK1 or TBK1 immunoblotting. Total substrate amounts were detected by Coomassie blue staining; note the shift in IRF3 protein upon phosphorylation. As controls, total immunoprecipitated protein was detected by FLAG immunoblot, whereas YopJ levels were confirmed by GFP immunoblot.
To confirm that YopJ acetylates mammalian TAK1, hTAK1 was expressed in the presence or absence of YopJ and then analyzed by tandem mass spectrometry. Similar to dTAK1, hTAK1 showed no acetylation when expressed alone; however, in the presence of YopJ, T184 and T187 within the activation loop of hTAK1 were acetylated (Fig. 4B). Because phosphorylation of T187 is critical for activation of hTAK1 (24–28), acetylation of this residue likely blocks phosphorylation and activation of the kinase. A number of other residues in the activation loop and beyond were also acetylated in the presence of YopJ, although definitive localization of some of the sites could not be unambiguously identified (Table S3).
Furthermore, we tested the YopJ-dependent acetylation of a number of mammalian MAP2 kinases downstream of TAK1 in mammalian systems. Previously, YopJ has been shown to acetylate MKK6 on ser207 and thr211, the activating phosphoacceptor sites in the activation loop (10). Consistent with published findings, YopJ blocked MKK6 phosphorylation on ser207 (Fig. S3A, Top). In addition to acetylating ser207 and thr211, YopJ also acetylates a neighboring lysine (lys210) (10). Although this lysine modification is dispensable for the inhibitory effect of YopJ, it provides a unique opportunity to detect YopJ-mediated acetylation by immunoblotting with an acetyl-lysine antibody (Fig. S3A, Middle). MKK4 and MKK7, the MAP2Ks that specifically activate JNK, also contain at least one lysine in their activation motif. Exploiting these nearby lysines revealed that YopJ acetylates both MKK4 and MKK7 (Fig. S3B). Consistent with this acetylation, YopJWT, blocked JNK phosphorylation by MKK7 overexpression (Fig. S3C). Although YopJ has been shown to interact with MKK4 or MKK7 (29, 30), these data provide direct evidence for YopJ-mediated acetylation and inhibition of these JNK-specific MAP2Ks. Our results argue that that YopJ is capable of modifying both a MAP3 kinase (i.e., TAK1) and MAP2 kinases (i.e., MKK4 and MKK7) in a single pathway (i.e., JNK) (Fig. 5). We therefore propose that this two-pronged attack underlies the potent inhibitory effects YopJ exerts on multiple signaling pathways.
Fig. 5.
YopJ inhibits MAP2 and MAP3 kinases. A model of YopJ-mediated inhibition, from our work and others, in both the Drosophila IMD (Left) and mammalian TNF/TLR (Right) signaling pathways. Proteins known to be acetylated by YopJ are marked with ‘Ac’ (red).
To date, previous work has failed to identify a kinase not affected by the acetyltransferase activity of YopJ. Our prior study on YopJ demonstrated that RIG-I–mediated IRF3 activation is impervious to YopJ-mediated inhibition (7). In this pathway, TBK1 is an essential IRF3 kinase (31–33). Thus, the ability of YopJ to interfere with hTAK1 and TBK1 kinase activities was compared directly. Consistent with the inability of YopJ to disrupt RIG-I signaling, it failed to inhibit TBK1 kinase activity while clearly blocking hTAK1 (Fig. 4C, row 1). Furthermore, phosphorylation of the TBK1 activation loop was not blocked by YopJ, whereas hTAK1 phosphorylation was inhibited (Fig. 4C, row 4). These results suggest that, despite the broad inhibitory profile of YopJ, this effector targets a subset of host kinases.
Discussion
Previous studies in mammalian systems have identified a number of targets of YopJ inhibitory activity. In one study, the YopJ homolog, YopP, was shown to inhibit mammalian TAK1; however, no mechanism was identified (11, 12). A number of in vitro studies identified the MAP2 kinase family as targets of YopJ-mediated serine/threonine acetylation (9, 10). Through these acetylations, it was argued that YopJ is capable of inhibiting both MAPK (ERK pathway in particular) and NF-κB signaling. The data presented here demonstrate that the MAP3 kinase TAK1 is also potently targeted by YopJ-mediated acetylation. This acetylation inhibits the critical autophosphorylation of TAK1, simultaneously blocking innate immune-induced NF-κB and JNK signaling of both Drosophila and mammals (Fig. 5).
We show that YopJ is able to inhibit Drosophila IMD but not Toll innate immune signaling by blocking the activity of dTAK1, after PGN stimulation or overexpression of dTAK1. However, upstream signaling events, such as IMD cleavage and ubiquitination, remain intact in the presence of YopJ. Instead, YopJ acetylates multiple serine and threonine residues in the dTAK1 activation loop, inhibiting autophosphorylation of this kinase. Furthermore, YopJ also inhibits the activation of mammalian TAK1. Because TAK1 belongs to the more divergent MAP3 kinases, these findings raise the possibility that YopJ may target other MAP3 kinases as well. However, YopJ did not inhibit the kinase TBK1 and, therefore, is not an inhibitor of all kinases. We also demonstrate that YopJ is also capable of acetylating and inhibiting the MAP2 kinases MKK4, MKK6, and MKK7. When put in context with previous work showing YopJ acetylation of mammalian IKK and MKKs (9, 10), our data provide evidence that YopJ is able to redundantly inhibit the JNK, p38, and NF-κB innate immune signaling pathways at critical MAP2 and MAP3 kinases (Fig. 5).
The precise mechanism of YopJ-mediated immune inhibition remains controversial. Previous work, from our group and others, suggests that YopJ acts as an ubiquitin protein protease, cleaving ubiquitin from conjugated substrates (7, 8), whereas experiments herein demonstrate that YopJ functions as a serine/threonine acetyl-transferase, acetylating critical residues on TAK1. Our earlier studies primarily concluded that YopJ-inhibited innate immune signaling upstream of IKK activation, consistent with our results here. We, and others, further argued that YopJ may have ubiquitin protease activity based solely on cotransfection studies. Here, we show in the context of ligand-induced innate immune signaling that Drosophila TAK1 is inhibited by YopJ, and that TAK1 is a direct target of YopJ-mediated acetylation, strongly arguing that this acetyltransferase activity is the critical activity responsible for the immunosuppressive function of YopJ. However, Zhou et al. present biochemical data that recombinant YopJ has ubiquitin protease activity (8). Therefore, it remains possible that YopJ has two enzymatic activities. Although we demonstrate that IMD ubiquitination is not perturbed in the presence of YopJ, IMD may not be a suitable substrate for YopJ-mediated deubiquitination.
Our approach, with the Drosophila model system, has many advantages over that previously described. First, this work was undertaken exclusively in live cells or animals stably expressing YopJ, an approach that has proven difficult within the context of mammalian cells because of the proapoptotic nature of YopJ (2). Using the inducibility of the Drosophila metallothionein promoter system, we developed stable cell lines, which provide reliable protein expression and overall consistency. Second, the proteins analyzed by MS/MS were produced and isolated from animal cells. The identification of TAK1 as the target for YopJ-mediated inhibition is also more in line with the observations regarding the ability of YopJ to inhibit both proinflammatory NF-κB signaling and MAPK pathways (1–4, 9, 10). In fact, while this work was under revision, Meinzer et al. reported that Yersina pseduotuberculosis YopJ acetylates TAK1, similar to our findings (34). Lastly, YopJ-mediated inhibition of hTAK1 in a mammalian context validates our model system approach. Targeted inactivation of TAK1 allows for a much broader and more efficient method of innate immune pathway inhibition. Perhaps in concert with inhibition of MAP2 kinases, YopJ is able to effectively inhibit immune pathways in both mammals and insects.
Materials and Methods
Cell Culture.
S2* cells were cultured in Schneiders media (Gibco) supplimented with 10% (vol/vol) FBS (Valley Biomedical), 1% (vol/vol) glutamine (Gibco), and 0.2% Pen/Strep (Gibco). Cells were treated with 1 μM 20-hydroxyecdysone (Sigma) for 24–30 h before stimulation with 100 ng/mL peptidoglycan (Invivogen) or 5 mM Spätzle (22).
RNA Analysis.
Total RNA was isolated with the TRIzol reagent (Invitrogen) as described (35) and expression of Diptericin and Rp49 was analyzed by Northern blot analysis followed by autoradiography.
Protein and Immunoprecipitation Assays.
Proteins were precipitated and analyzed as described (7, 16). See SI Materials and Methods for more details.
Kinase Assays.
Kinases were assayed as described (16, 35, 36), with modifications for cold kinase assays (see SI Materials and Methods for more details). GST-IRF3 aa380-427 was used as a substrate for TBK1 as described (32).
Peptide Identification by Tandem Mass Spectrometry.
See SI Materials and Methods for more details.
Supplementary Material
Acknowledgments
N.P. was supported by New England Regional Center of Excellence Grant U54AI057159. N.S. and L.S. were supported by National Institutes of Health Grants AI060025 and AI053809, and 1R01AI079198, respectively. N.S. was also supported by the Ellison Medical Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008203109/-/DCSupplemental.
References
- 1.Monack DM, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monack DM, Mecsas J, Bouley D, Falkow S. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J Exp Med. 1998;188:2127–2137. doi: 10.1084/jem.188.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer LE, Hobbie S, Galán JE, Bliska JB. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 4.Palmer LE, Pancetti AR, Greenberg S, Bliska JB. YopJ of Yersinia spp. is sufficient to cause downregulation of multiple mitogen-activated protein kinases in eukaryotic cells. Infect Immun. 1999;67:708–716. doi: 10.1128/iai.67.2.708-716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schesser K, et al. The yopJ locus is required for Yersinia-mediated inhibition of NF-kappaB activation and cytokine expression: YopJ contains a eukaryotic SH2-like domain that is essential for its repressive activity. Mol Microbiol. 1998;28:1067–1079. doi: 10.1046/j.1365-2958.1998.00851.x. [DOI] [PubMed] [Google Scholar]
- 6.Orth K, et al. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 7.Sweet CR, Conlon J, Golenbock DT, Goguen J, Silverman N. YopJ targets TRAF proteins to inhibit TLR-mediated NF-kappaB, MAPK and IRF3 signal transduction. Cell Microbiol. 2007;9:2700–2715. doi: 10.1111/j.1462-5822.2007.00990.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, et al. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J Exp Med. 2005;202:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 11.Thiefes A, et al. The Yersinia enterocolitica effector YopP inhibits host cell signalling by inactivating the protein kinase TAK1 in the IL-1 signalling pathway. EMBO Rep. 2006;7:838–844. doi: 10.1038/sj.embor.7400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haase R, Richter K, Pfaffinger G, Courtois G, Ruckdeschel K. Yersinia outer protein P suppresses TGF-beta-activated kinase-1 activity to impair innate immune signaling in Yersinia enterocolitica-infected cells. J Immunol. 2005;175:8209–8217. doi: 10.4049/jimmunol.175.12.8209. [DOI] [PubMed] [Google Scholar]
- 13.Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci USA. 2002;99:12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko T, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 16.Paquette N, et al. Caspase-mediated cleavage, IAP binding, and ubiquitination: Linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman N, et al. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- 20.Ertürk-Hasdemir D, et al. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 22.Weber AN, et al. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky MH, et al. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 24.Kishimoto K, Matsumoto K, Ninomiya-Tsuji J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J Biol Chem. 2000;275:7359–7364. doi: 10.1074/jbc.275.10.7359. [DOI] [PubMed] [Google Scholar]
- 25.Yu Y, et al. Phosphorylation of Thr-178 and Thr-184 in the TAK1 T-loop is required for interleukin (IL)-1-mediated optimal NFkappaB and AP-1 activation as well as IL-6 gene expression. J Biol Chem. 2008;283:24497–24505. doi: 10.1074/jbc.M802825200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prickett TD, et al. TAB4 stimulates TAK1-TAB1 phosphorylation and binds polyubiquitin to direct signaling to NF-kappaB. J Biol Chem. 2008;283:19245–19254. doi: 10.1074/jbc.M800943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiao B, Padilla SR, Benya PD. Transforming growth factor (TGF)-beta-activated kinase 1 mimics and mediates TGF-beta-induced stimulation of type II collagen synthesis in chondrocytes independent of Col2a1 transcription and Smad3 signaling. J Biol Chem. 2005;280:17562–17571. doi: 10.1074/jbc.M500646200. [DOI] [PubMed] [Google Scholar]
- 28.Singhirunnusorn P, Suzuki S, Kawasaki N, Saiki I, Sakurai H. Critical roles of threonine 187 phosphorylation in cellular stress-induced rapid and transient activation of transforming growth factor-beta-activated kinase 1 (TAK1) in a signaling complex containing TAK1-binding protein TAB1 and TAB2. J Biol Chem. 2005;280:7359–7368. doi: 10.1074/jbc.M407537200. [DOI] [PubMed] [Google Scholar]
- 29.Orth K, et al. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- 30.Du F, Galán JE. Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLoS Pathog. 2009;5:e1000595. doi: 10.1371/journal.ppat.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 32.McWhirter SM, et al. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinzer U, et al. Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates Caspase-1 to induce intestinal barrier dysfunction. Cell Host Microbe. 2012;11:337–351. doi: 10.1016/j.chom.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Silverman N, et al. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.